Journal Name: International Journal of Cancer and Treatment
Article Type: Research
Received date: 27 August, 2018
Accepted date: 30 August, 2018
Published date: 03 September, 2018
Citation: Kassem NM, Sharaf S, Aziz AA, Mohsen M, Kassem HA, et al (2018) Towards Validation of Targeted Next-Generation Sequencing on Formalin Fixed Paraffin Embedded Colorectal Cancer Tissues in Egyptian Population: A Pilot Study with Feasibility and Challenges. Vol: 1, Issu: 1 (20-29).
Copyright: © 2018 Kassem NM, et al. This is an openaccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Colorectal cancer (CRC) has been identified as the third most common cancer worldwide. Gene mutation and defective cell regulation are important processes in the development of CRC. Assessment of genetic mutations is an essential element in the modern era of personalized cancer treatment.
Purpose: We performed a pilot study identifying the frequencies of genetic changes in six frequently mutated genes: KRAS, NRAS, BRAF, PIK3CA, PTEN and TP53 in Egyptian CRC patients. The secondary objective was to validate the next-generation sequencing (NGS) technology and to develop a workflow process in the clinical setting.
Methodology: This study investigates the mutations in these 6 genes using targeted NGS. The study included 14 archived Formalin-fixed paraffin-embedded (FFPE) tumor specimens in addition to 11 external quality controls (EQC).
Results: The median age of our CRC patients was 60.5 years, 21.4% of patients were ≤ 40 years old, females 57.1% and males 42.9% with M/F ratio 0.75:1. The mutation frequencies of KRAS, BRAF, PIK3CA, PTEN and TP53 genes in Egyptian CRC patients were 35.7%, 14.3%, 21.4%, 7.1% & 71.4% respectively, with no mutations detected in NRAS gene which may be due to the limited study population.
Conclusion: Technical validity and clinical efficacy are the two major arguments in the analysis of NGS data. The major benefit of NGS in comparison to more traditional methods is its ability to study multiple genes at once. Implementation of targeted NGS in clinical settings allows for a trustworthy description of the most common mutations, which can direct therapeutic decisions for CRC patients.
Keywords
Colorectal cancer, NGS, Genetic mutations, Targeted therapy.
Abstract
Background: Colorectal cancer (CRC) has been identified as the third most common cancer worldwide. Gene mutation and defective cell regulation are important processes in the development of CRC. Assessment of genetic mutations is an essential element in the modern era of personalized cancer treatment.
Purpose: We performed a pilot study identifying the frequencies of genetic changes in six frequently mutated genes: KRAS, NRAS, BRAF, PIK3CA, PTEN and TP53 in Egyptian CRC patients. The secondary objective was to validate the next-generation sequencing (NGS) technology and to develop a workflow process in the clinical setting.
Methodology: This study investigates the mutations in these 6 genes using targeted NGS. The study included 14 archived Formalin-fixed paraffin-embedded (FFPE) tumor specimens in addition to 11 external quality controls (EQC).
Results: The median age of our CRC patients was 60.5 years, 21.4% of patients were ≤ 40 years old, females 57.1% and males 42.9% with M/F ratio 0.75:1. The mutation frequencies of KRAS, BRAF, PIK3CA, PTEN and TP53 genes in Egyptian CRC patients were 35.7%, 14.3%, 21.4%, 7.1% & 71.4% respectively, with no mutations detected in NRAS gene which may be due to the limited study population.
Conclusion: Technical validity and clinical efficacy are the two major arguments in the analysis of NGS data. The major benefit of NGS in comparison to more traditional methods is its ability to study multiple genes at once. Implementation of targeted NGS in clinical settings allows for a trustworthy description of the most common mutations, which can direct therapeutic decisions for CRC patients.
Keywords
Colorectal cancer, NGS, Genetic mutations, Targeted therapy.
Introduction
Colorectal cancer has been determined as the 3rd most common cancer worldwide [1]. It represents 7.4% of all diagnosed cancer cases in the area of Middle East and North Africa [2]. In Egypt, total number of 41919 primary malignant tumors coded data was collected from the NCI computer database including all records documented during the 12-year period from 2000-2011. It represents a hospital-based pathology registry based on precise tissue diagnosis using advanced ancillary techniques and following the recent WHO classification of tumors. This Egyptian registry revealed that the prevalence of GIT tumors was 18.54% from total malignancies. Colon cancer prevalence was 15.3% from total GIT tumors percentage and 2.83% from total malignancies. The prevalence of rectal cancer was 18.5% from total GIT tumors percentage and 3.43% from total malignancies. There was a slight women predilection over men patients (51.2% and 48.8% respectively). Majority of primary neoplastic cases were of epithelial nature, with the majority were carcinomas (97.8%). Adenocarcinoma (NOS) was the commonest type diagnosed (72.7%) [3]. Multiple genetic events accumulate during the progression of colorectal carcinogenesis. Detection of genetic mutations is a meaningful factor in the era of personalized cancer therapy. In CRC, these genetic events include microsatellite instability and chromosomal instability which has limited predictive or prognostic value and do not influence treatment decision in the metastatic patients. Mutation in KRAS is the most crucial widely used molecular test in the CRC metastatic settings due to presence of this mutation can predict lack of benefit from EGFRtargeted antibodies [4]. Also, the patients with mutations in both BRAF and the PIK3CA have shown negative response to treatment with EGFR inhibitors [5]. KRAS mutation is found in 17%-25% of all cancers and 35%-45% of CRCs [6]. Mutations of KRAS are highly frequent in the codons: 12; 13; 59; 61 & 146 [7,8]. NRAS is the 2nd most important RAS protein involved in CRC, after KRAS [9]. NRAS is generally observed mutated in 15% of all cancers and 5% of all CRCs [9,10]. NRAS mutations previously reported to have a strong association with KRAS-wild type and are generally found to be muted exclusively with KRAS mutations [11,12]. BRAF gene mutations are persistent in 15% of all cancers, and the V600E mutation is present in 10% of all CRCs [13,14]. A significant higher BRAF mutations frequency had been demonstrated in females and in the proximal colon [15-18]. Furthermore, KRAS and BRAF mutations are observed to be nearly mutually exclusive of each other [19,20]. PIK3CA gene is implicated in PI3K/ Akt pathway and is observed mutated in ~ 10%-20% of CRCs where approximately 80% mutations are detected in two hotspot areas in exons 9 and 20 [21]. These PIK3CA mutations are more frequent in the proximal colon [22,23]. PTEN gene is a tumor suppressor gene located at chromosome 10q23.31. PTEN main function is to block the PI3K pathway by dephosphorylating phosphatidylinositol (PI) 3, 4, 5-triphosphate into PI-4, 5-bisphosphate thus opposing PI3K function. In CRC, PTEN is changed by genetic & epigenetic mechanisms; typically, mutations, promoter hypermethylation and 10q23 LOH, which lead to the biallelic inactivation of the protein in 20-30% of cases [24]. TP53 gene which is tumor suppressor gene regulates DNA repair mechanism and apoptosis. The TP53 mutation is observed in nearly 50% CRCs, with the most frequently observed mutations between exons 5 and 8 at codons 175, 213, 245, 248, 273, and 282 [25,26]. Currently, molecular-based targeted therapy is a standard approach in selected patients such as those with unresected metastatic CRC disease being eligible for inhibitors of VEGF and EGFR. As reported by the 2016 ESMO guidelines, targeted therapy can be used mutually with cytotoxic regimens for unresected metastatic CRC patients [27].
Subjects and Methods
Study Objectives
The present study was exploratory in nature. The first intent was to demonstrate the genetic changes in Egyptian CRC patients, so as to expand the range of potential mutational targets with therapies that were commercially available. This study includes six frequently mutated genes: KRAS, NRAS, PIK3CA, BRAF, PTEN and TP53. Mutations in these 6 genes have been broadly investigated because of the high prevalence observed in CRC patients, and this research explores these mutations using targeted NGS in Egyptian population. The second intent was to validate the NGS methodology and to establish a workflow process; this process should occur in a short enough timeframe in order for patients to gain benefit from this additional information in developing a cancer patient treatment plan.
Sample collection and quality controls
Fourteen archived FFPE tumor specimens from 14 colorectal cancer patients treated at Kasr Al Ainy Clinical Oncology department - Cairo University were randomly chosen from the Pathology department existing registry. This study was approved by Kasr Al Ainy Clinical Oncology department Institutional Review Board (IRB).
Validation was done on 2 steps; the first one was performed using FFPE DNA samples, cell lines besides commercially available reference DNA samples provided by Illumina. The samples will have representative mutation within each gene (KRAS and NRAS) along with different codons 12; 13; 59; 61; 117 and 146. The study will determine if mutations can be called accurately. DNA obtained from FFPE specimens (Panel C: C27, C33, C34, C35 & C36) will also be tested to determine accuracy of the assay. This study will also determine if operator(s) are trained to undertake testing. DNA extracted from cell lines (RPMI 8226, MDAMB- 231 & HL-60) were used for the proficiency testing. The version number of the software was documented at the start of all testing for every run and tested to determine if expected variant calls are present in the.vcf file. The annotation software was also tested using the.vcf file as input and the output including the expected corresponding gene, exon and amino acid information. The second validation step was performed by participation in External Quality Assessment Schemes 2016-2017 (UK-NEQAS) for molecular genetic analysis of (KRAS, NRAS BRAF and PIK3CA) genes in CRC round 2.
DNA extraction from patients’ samples & quality controls
Genomic DNA was extracted from formalin fixed paraffin embedded (FFPE) tissues with the QIAamp DNA Mini kit (Qiagen, USA) as reported by the manufacturer and was eluted in 60 μL volume. The extracted DNA samples were measured utilizing the Qubit dsDNA High Sensitivity (HS) assay kit (Life Technologies-Fisher Scientific, USA) and samples with DNA concentration lower than 10 ng/ μl were excluded. A quantitative PCR (qPCR) reaction was performed to conclude the amplifiability of the FFPEextracted gDNA samples. By analyzing the amplifiability of FFPE DNA in relation to that of the QCT non-FFPE reference gDNA, a ΔCq value can be calculated for each sample and used to anticipate its efficiency in the library preparation assay. Samples were considered passing quality control if the gDNA samples giving a delta Cq value of less than 4.
Illumina control samples were provided as extracted FFPE DNA, cell lines along with commercially available reference DNA samples; while UK-NEQAS samples were provided as rolled sections of paraffin embedded tumour tissue for each case to be extracted as described above.
Library preparation from patients’ samples & quality controls
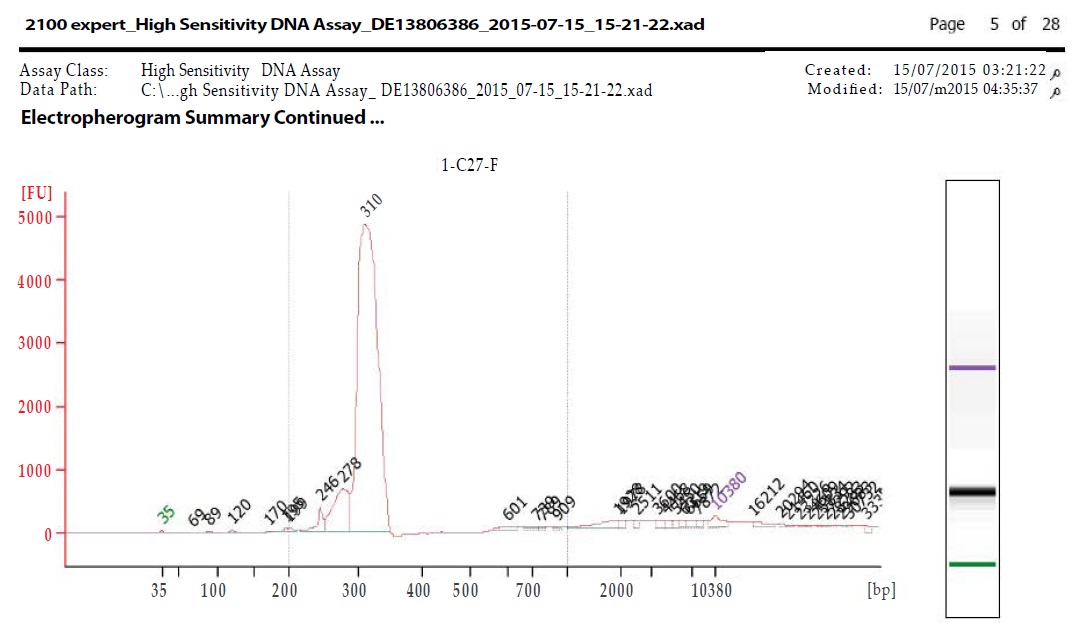
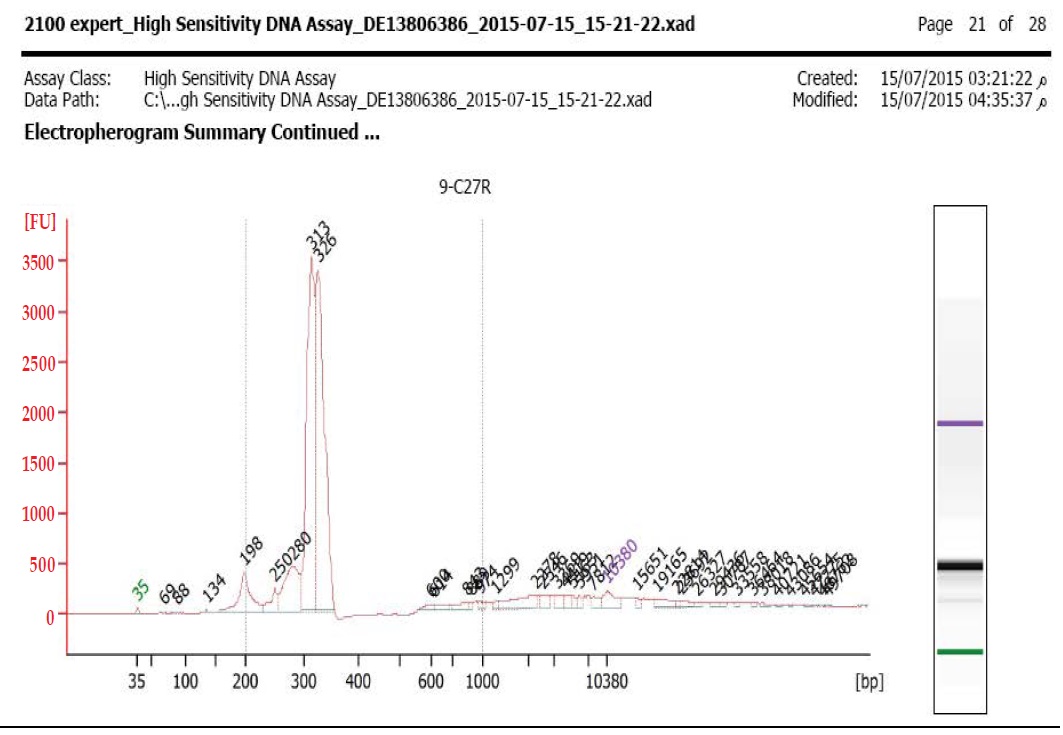
Libraries were prepared using TruSight Tumor 26 kit according to manufacturer’s Protocol (Illumina, Inc., San Diego, CA). In brief, an oligo pool was hybridized to the genomic DNA sample. After the unbound oligos removal, target areas of interest flanked by sequences needed for amplification were generated by extension and ligation; then libraries were further PCR amplified. Library quality was checked out by 2100 Bioanalyzer utilizing the Bioanalyzer High Sensitivity (HS) reagents & Chips (Agilent Technologies, Santa Clara, California). Libraries were estimated to pass QC as long as they had an average peak size between ~300-330 bp as demonstrated in Figure 1a,1b. Before sequencing, the libraries were normalized following the manufacturer’s protocol and equal volumes were pooled to constitute the terminal sequencing library. The TruSight Tumor Sequencing Panel achieves limits of detection below 5%variant allele frequency across 174 amplicons with 1,000x minimum coverage of each amplicon.
Figure 1a: Bioanalyzer for C27 forward primer.
Figure 1b: Bioanalyzer for C27 reverse primer.
Sequencing and Data Analysis
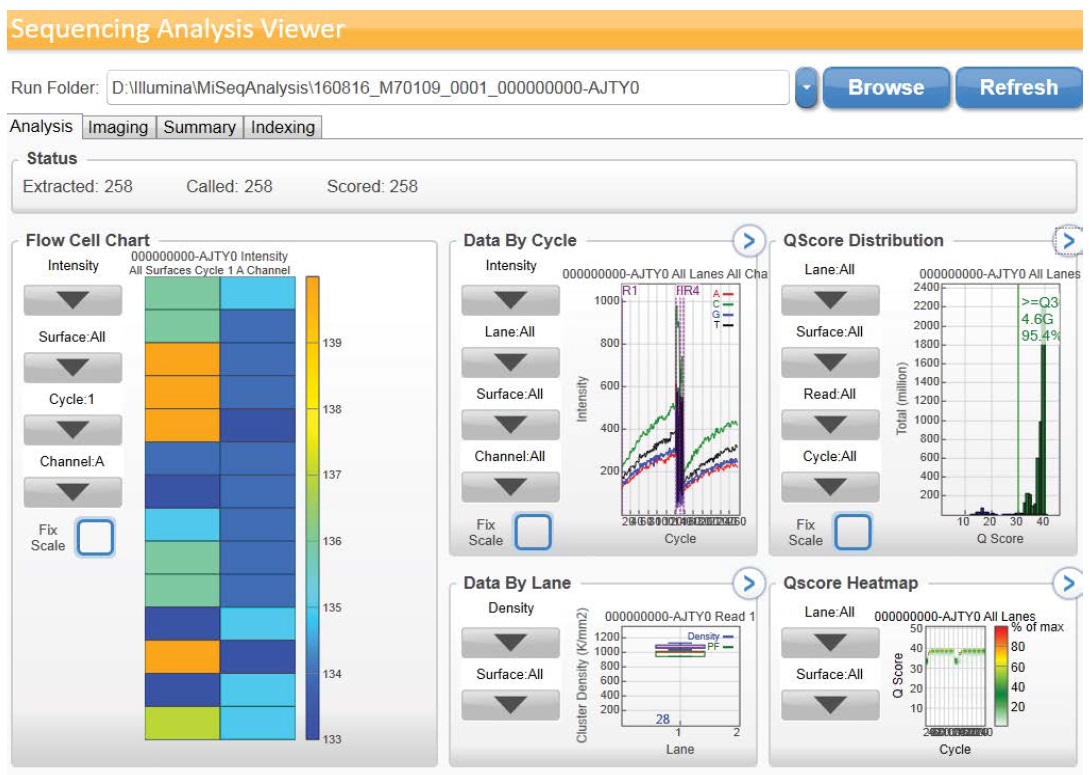
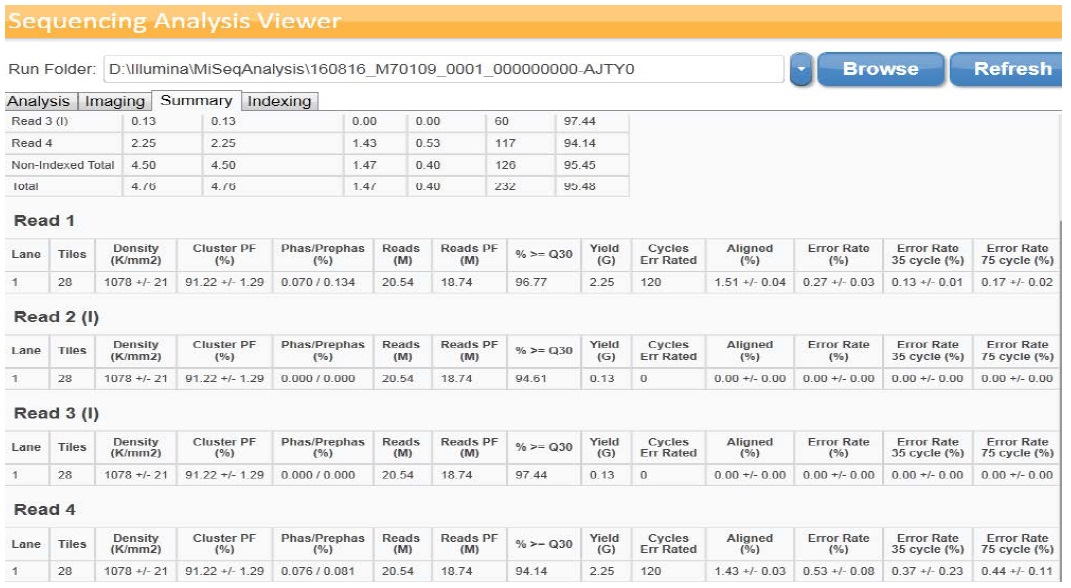
Sequencing was accomplished utilizing MiSeqDx device (llumina) with a 2 × 150 bp read length and total time of ~ 24 hours which involve cluster generation, sequencing and base calling on the MiSeqDx system. Specifications based on Illumina PhiX control library support cluster densities between 865-965 k/mm2 clusters passing filter for v2 chemistry. The quality scores > 80% bases higher than Q30 at 2 x 150 bp as demonstrated in Figure 2a,2b. Image processing and VCF file generation were further analyzed, we then annotated the variants using VariantStudio™ software version 2.2.1.175. Noncoding regions and synonymous variants were filtered out. Mutations with low depth, which indicate ≤ 50x depths, were filtered out. In addition, mutations with ≤ 5% variant allele frequency were filtered out. Variant quality which is one parameter of the variant call format (VCF) < 80% was filtered out. The VariantStudio™ software provides the numerical identifier for the variant in COSMIC database, if the genomic variant position overlaps a variant listed in COSMIC. The COSMIC ID connects to the COSMIC page associated with the identifier to provide information on whether that variant has been observed before in any tumor variety or a novel one.
Figure 2a: Graphical data obtained by illumina Sequencing Analysis Viewer.
Figure 2b: Summary of data obtained by illumina Sequencing Analysis Viewer.
Results
Patients’ characteristics
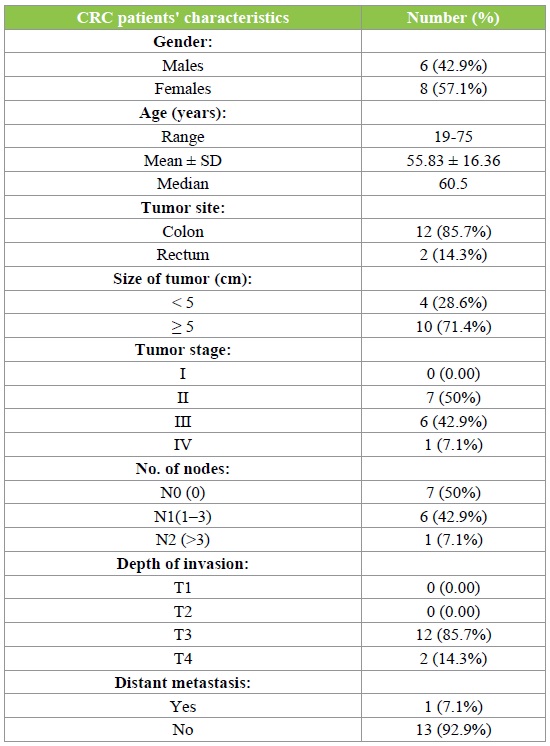
Fourteen CRC patients were incorporated in our work besides controls. Patients’ characteristics are given in Table 1. The median age of our CRC patients was 60.5 years (range, 19-75 years), 3 (21.4%) patients were ≤ 40 years old. Among the studied cases, CRC was more frequently observed in females (57.1%) than males (42.9%) with M/F ratio 0.75:1. Thirteen patients (92.9%) were invasive adenocarcinoma grade II & only one patient (7.1%) was grade III. Primary tumors were higher in colon (85.7%) than rectum (14.3%). The size of tumor was reported to be ≥ 5 cm in 10 (71.4%) patients. A higher number of patients were noted to have tumor stage II (50%), followed by stage III (42.9%) and stage IV (7.1%). The N0, N1 & N2 stages of lymph node invasion were noted to have the similar percentages. However, a noteworthy variation was noticed in the invasion depth of tumor in the layers of colon/rectum, where T3 represented 85.7% of all cases, followed by 14.3 % cases in T4 and no case observed in T1or T2. Finally, presence of distant metastasis was noticed in 3 patients (7.1%) of our study population.
Table 1: CRC patients' characteristics.
NGS patients’ results
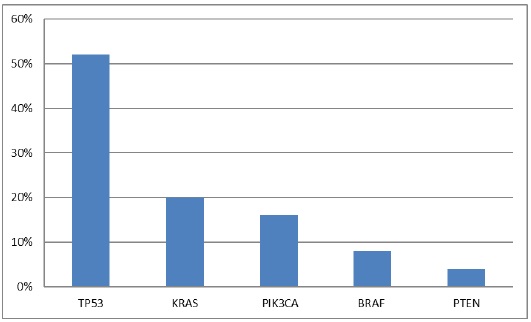
To determine somatic mutations, we performed the targeted NGS on 14 CRC patients` samples. We identified 25 mutations which were missense mutations. These mutations list in all patients is reported in Table 2. Two (14.3%) of 14 patients had no mutation detected; 4 (28.6%), 5 (35.7%), 2 (14.3%), and 1 (7.1%) patients had 1, 2, 3, and 5 somatic mutations, respectively. The frequency of each somatic mutation is demonstrated in Figure 3, the most repeatedly mutated gene was TP53 (13 mutations), followed by KRAS (5 mutations), PIK3CA (4 mutations), BRAF (2 mutations) and PTEN the lowest with only one mutation, while no mutations in NRAS gene.
Table 2: Patients' mutational data.
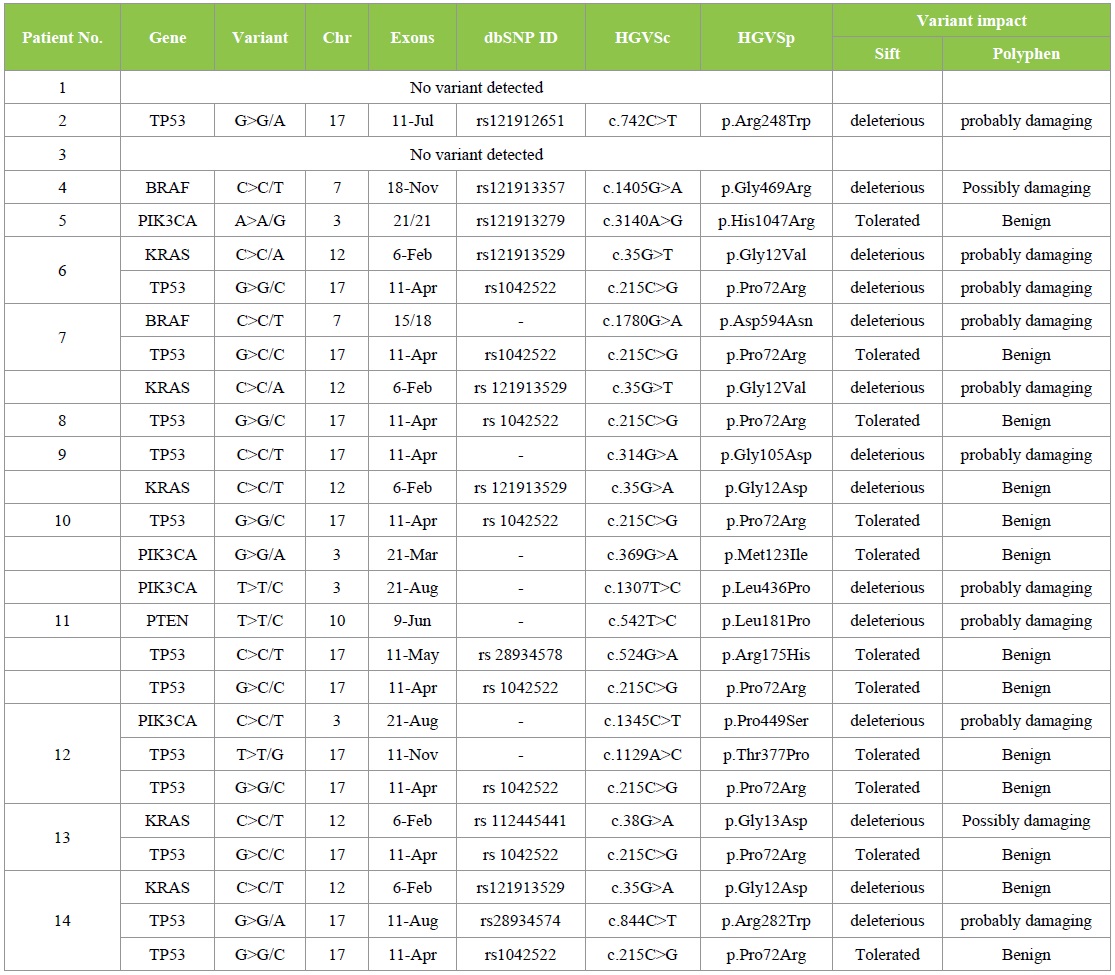
Figure 3: Frequency of somatic mutations in 14 Egyptian CRC cases.
TP53 mutations was noticed in 10 out 14 (71.4%) of our CRC patients, with total of 13 mutations. Three patients each harbor two types of TP53 mutations: R175H, P72R; T377P, P72R & R282W, P72R, while the rest harbored only one TP53 mutation. TP53 mutations were evenly detected in both genders. The six codons for TP53 mutations were 72, 105, 175, 248, 282, and 377; accounted for (8/13; 61.5%) in the codon 72 and (1/13; 7.7%) for each one of the rests. The most frequent TP53 mutation was P72R that individually account for eight cases and occur as a result of substitution at c.215C > G. Furthermore, five TP53 mutations G105D, R175H, R248W, R282W and T377P were noted in one patient each. These five TP53 mutations occur as a result of substitution at c.314G > A, c.524G > A, c.742C > T, c.844C > T and c.1129A > C, respectively and together constitute 5/13 (38.5%) of all TP53 mutations.
KRAS mutations were noted in 5/14 (35.7%) patients, afford to total of 5 mutations. A higher prevalence of KRAS mutations was noticed in female patients (3/5, 60%) than male patients (2/5, 40%). Codon 12 & 13 constituted (4/5, 80%) & (1/5, 20%) of all KRAS mutations respectively. These mutations were substitution of glycine with aspartate or valine on codon 12 and substitution of glycine with aspartate on codon 13. These 3 mutations G12D, G12V and G13D occur as a result of substitution at c.35G > A; c.35G > T & c.38G > A and have been detected in 2/5 (40%), 2/5 (40%), and 1/5 (20%) patients, respectively.
Mutated PIK3CA was noticed in 3/14 (21.4%) patients. Each patient harbored only one PIK3CA mutation except for one patient with two PIK3CA mutations, M123I and L436P. A higher prevalence of PIK3CA mutations was noticed in female patients (2/3, 66.7%) than male patients (1/3, 33.3%). The four mutations observed were H1047R, P449S, M123I and L436P which occur as a result of substitution at c.3140A > G, c.1345C > T, c.369G > A and c.1307T > C, respectively.
BRAF mutations were determined in 2/14 (14.3%) patients. A single BRAF mutation was noticed in every patient. In our CRC patients, BRAF mutations were evenly observed in both genders. In our study, BRAF mutations were G469R, which occurs as a result of substitution at c.1405G > A and D594N which occurs as a result of substitution at c.1780G > A.
Mutated PTEN was determined in 1/14 (7.1%) patient. In our study, PTEN mutation occurred in only one male patient and was L181P, as a result of substitution at c.542T > C.
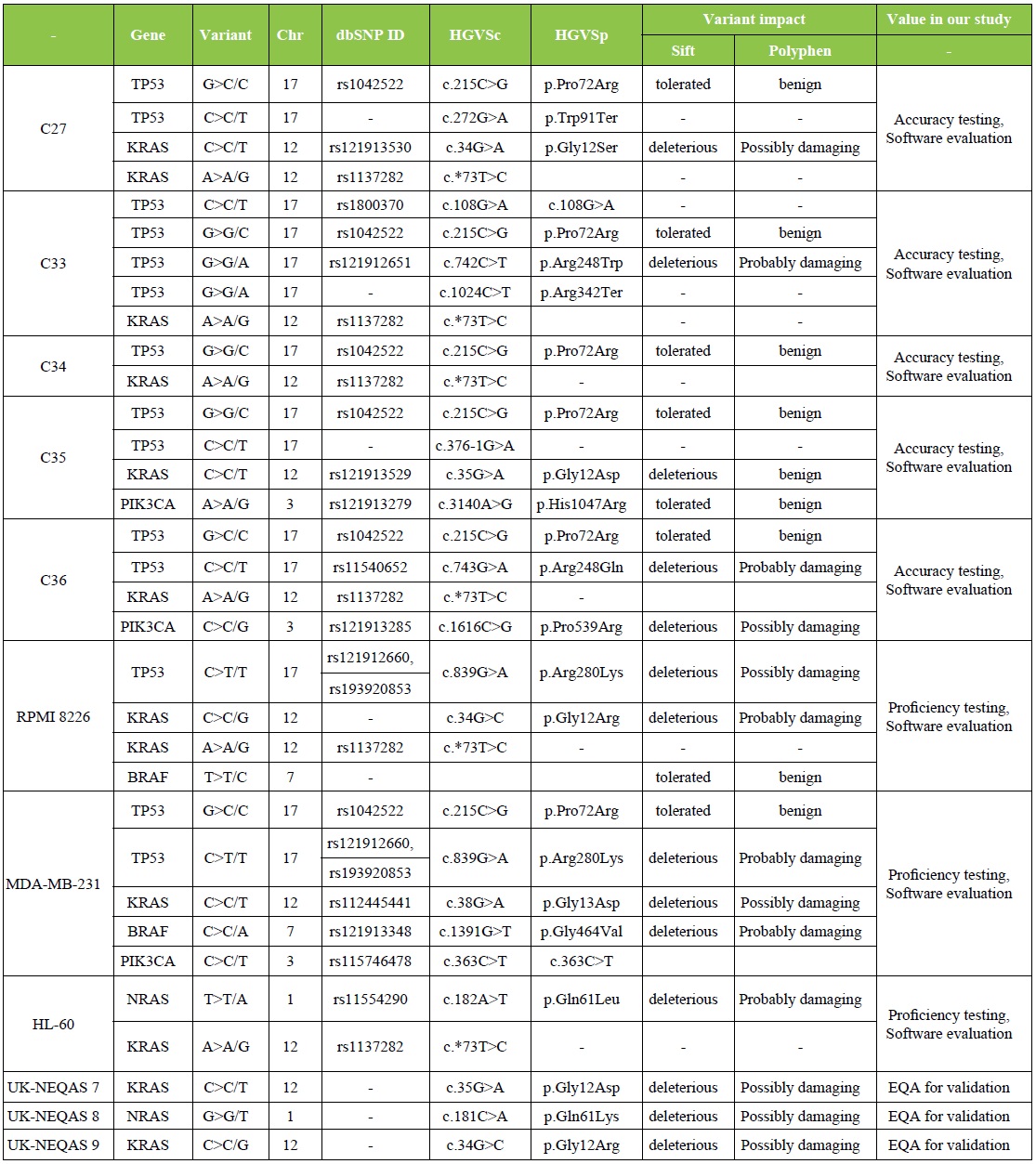
External Quality Control NGS results
Validation of NGS methodology was done utilizing 8 illumina controls and 3 UK-NEQAS samples. The list of mutations detected by our Lab. in all external quality controls is demonstrated in Table 3. Our Molecular Lab. had successfully participated in External Quality Assessment Schemes 2016-2017 (UK-NEQAS) for molecular genetic analysis of KRAS, NRAS BRAF and PIK3CA in CRC round 2.
Table 3: External Quality Control Sequencing results.
Discussion
In the coming era of personalized cancer treatment, it is essential to have exact genetic information of each individual cancer. An extremely large number of genetic information even now direct treatment decisions in ordinary clinical practice. We have established and validated a targeted NGS technology to provide global genetic information for each patient. The main privilege of the targeted NGS is that data in regard to multiple genes and multiple genetic variations is generated with a single experiment. The sequencing device providing genetic mutational data requires fewer DNA amounts and cost compared with testing individual genetic alteration by PCR or FISH which are frequently used in the daily practice. Moreover, superior sensitivity over Sanger sequencing can be obtained by increasing coverage depth, especially in cases with low tumor purity [4].
In this pilot study, CRC was found to be higher in women (57.1%) than men (42.9%) which corresponds to the results previously reported in Egypt [3] while contradictory to the results reported by studies worldwide [1,28]. The cancer was observed predominantly in colon (85.7%) over rectum (14.3%) which is comparatively with that previously reported [29,30]. The median age of our CRC patients was 60.5 years (range, 19-75 years), and 3 (21.4%) patients were ≤ 40 years old. A detectable variation between the predominance of CRC in the East and West can be observed in the average age of diagnosis. In the US and European Union, only about 2-8% of cases occur in individuals under 40 years of age, whereas Egypt, Saudi Arabia, the Philippines, and Iran show rates of 38%, 21%, 17%, and 15-35%, respectively for this same age group [31].
KRAS mutational status is critical when considering anti- EGFR treatment for metastatic CRC patients. Even a number of methodologies are presently used to test archived FFPE samples for KRAS mutations in laboratory practice; the NGS feasibility in this regard has not been well established. In FFPE tissues, various studies have demonstrated high concordance rates between results obtained from 454 or Illumina NGS platforms and standard assays for KRAS mutation [32]. In our study, KRAS mutation was noted in 35.7% of CRC patients. This is comparatively with the average level of KRAS mutations (35%-45%) found in CRCs [6] and the mutation proportions observed in Serbia, Turkey, and Saudi Arabia (32%-34.7%); lower than the mutation proportions observed in Slovenia, Iraq, and Spain (46.2%-48%); and higher than the mutation proportions observed in Morocco and Greece, and previously reported proportion in India (23.5%-29.3%) [19,20,33]. A higher prevalence of KRAS mutations was noticed in female patients (60%) than male patients (40%). This corresponds to the results previously reported in Egypt [34]. However, in other reports an even distribution of KRAS mutations has been described in both genders [23,35]. The frequency of KRAS mutations at codons 12 and 13 were found to be 80% and 20%, respectively, which is comparatively to the previous Egyptian study which report that 68% of the KRAS mutations were in codon 12 and 23% in codon 13 [36].
The three mutations observed in our study, Gly12Asp, Gly12Val, and Gly13Asp (40%, 40%, and 20% of all KRAS mutations, respectively), are similar to the earlier reported mutation by Tong et al., who found that the most prevalent mutation was glycine to aspartate at codon 12 (G12D), which estimated as 37.5% of all mutations. Mutation from glycine to valine (G12V) was the 2nd most common of all specified mutations (20.1%; 135 of 672). Mutation from glycine to aspartate at codon 13 (G13D) accounted for 19.0% (128 of 672) of specified mutations [37]. A mutant form of TP53 was detected in 71.4% CRC patients. This result is comparatively with the frequencies reported by Austria, the United Kingdom, Scotland, Japan, the United States, China, France, Spain, Poland, and Norway (45%–84%) [29] which are significantly higher than the pooled frequency rates encountered in Middle Eastern (38.4%) and Asian countries (40.1%) [5]. The TP53 mutations were evenly observed in male besides female CRC patients. This in contrast to earlier literature, TP53 mutations were more common in male patients [38,39]. The six codons for TP53 mutations were 72, 105, 175, 248, 282, and 377; individually accounted for (61.5%) on codon 72 and (7.7%) for each one of the rest. El- Serafi et al., reported that TP53 mutation detected in codon 72 in (12/29 patients; 41.4%) in Egyptian CRC patients [40].
This mutational hotspot on exon 4 (codon 72) was not described previously in CRCs from Western countries. However, codon 72 polymorphism (arginine-proline) have been described in other solid tumors [41]. It is interesting to notice that this polymorphism in codon 72 has been demonstrated in other solid tumors in the Egyptian population, including bilharzial bladder cancer [42] and hepatocellular carcinoma [43], suggesting that there are specific environmental or racial contributions in our population. The frequency of mutations in codons 175, 248, 282, and 377 can be compared to previous reports (9%, 9% and 5% respectively) [44]. The mutation prevalence of PIK3CA in this work was found to be 21.4%. Very few studies describe the prevalence of PIK3CA mutations in an Arab population. Previous study [45] reported a PIK3CA frequency rate of 12.2% in a Saudi population, which is comparatively with the frequency noted in our Arab patients’ population (13%) and the Western patients cohort (12.1%). However, the frequency proportion of PIK3CA mutations in Asian countries (8.5%) appears to be lower in comparison with Middle Eastern and Western countries. This difference could be due to either environmental or genetic factors [5]. PIK3CA mutations were higher in women (66.7%) than men (33.3%). Also, previous studies have either reported more PIK3CA mutations in females [46] or an even distribution between males and females [23].
We identified BRAF mutations in 14.3% CRC patients. Previous study reported the frequency of BRAF mutation as 2.5% in 770 Saudi patients [47], which is lower than the Western cohort and the estimated frequency in Western countries (9.2%) but similar to the frequency reported in Asian (4.9%) and Middle Eastern countries (4.6%). This dissimilarity in BRAF mutational frequency may be due to the use of different methods & assays to evaluate for these mutations. In CRC patients, BRAF mutations were evenly detected in men and women. Interestingly, the Western population-based studies reported that BRAF-mutant CRC was significantly more likely to occur in females [48]; however, in other study, reported that there was no meaningful association between the BRAF mutation and gender [5]. In our CRC patients PTEN is changed through a mixed genetic-epigenetic mechanism; typically mutations, 10q23 LOH and promoter hypermethylation, which leads to the biallelic inactivation of the protein in 20-30% of cases [24]. In our study, mutated PTEN was identified in only one male CRC patient. This mutation was L181P, which occurs as a result of substitution at c.542T > C.
As NGS is an arising technology, there were expected challenges encountered during the conduction of this study. In this exploratory pilot study, we noticed several obstacles in performing NGS-based targeted therapy testing. First, the cost of NGS kit is a challenging issue as it is not covered by Egyptian health insurance. Second, technical challenges affecting NGS pathology FFPE samples are either low extracted DNA input or quantity (< 10 ng/μl) or low DNA quality (delta Cq > 4) or even both. This is mostly due to formalin used that should be freshly prepared twice per week and handled with care in a concentration of 10% neutral buffered in suitable containers. Usually in consideration of getting an NGS run of 8 samples, we could try DNA extraction more than double this number. Finally, limited access to targeted medications was the most prevalent cause that patients could not receive the targeted therapy. As a result, we should do vigorous efforts to make NGS technologies available for incorporation into routine practice together with targeted therapy availability for each cancer patient. Also, this work has a few limitations as a small group of patients were joined for evaluating the feasibility of targeted NGS technology.
Conclusion
Targeted NGS technology was able to support broad genetic alteration data in CRCs and can be used in clinical setting. Technical validity and clinical efficacy are the two major arguments in the analysis of NGS data. The major benefit of NGS in comparison to more traditional methods is its ability to study multiple genes at once. If the expenses are estimated per sequenced base, NGS is competitive to the other techniques. Implementation of targeted NGS in clinical settings allows for a trustworthy description of the most frequent mutations, which can direct therapeutic decisions for CRC patients.
Acknowledgement
We deeply acknowledge the scientific & technical support offered by Amgen Co., AGBL Co. Also, the great support from: Prof. Y. Abdel Kader, Prof. H. Abdel Azim, Prof. E. Hamada & Prof. W. Metenawy. We also pay homage to the soul of Prof. H. Atef. They all participated in the success of this pilot study. We are also thankful for the oncology dep. molecular lab. & bioinformatics teams.
Funding
This study was funded by Kasr Al Ainy Oncology dep., Cairo University & Amgen Co.
Conflicts of interest
Authors declared no conflicts of interest.
Ethical approval
All procedures performed in the study involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments (GCP guidelines) or comparable ethical standards.
Informed consent was obtained from all individual participants included in the study.
There is no references