Journal Name: Journal of Applied Microbiological Research
Article Type: Research
Received date: 19 October, 2020
Accepted date: 04 December, 2020
Published date: 04 January, 2021
Citation: Cao Z, Chen Q, Aizezi M, Li C, Zhu M, et al. (2021) A Reported Antarctic Environmental Microorganism Isolated from Mongolian Plateau. J Appl Microb Res. Vol: 4 Issu: 1 (01-06).
Copyright: © 2021 Cao Z et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Herein, we report a novel Carnobacterium-like organism, CS13T, isolated from Mongolian Plateau is 100% related to the reported Antarctic environmental microorganism Carnobacterium antarticum sp. CP1. The CS13T was isolated from 78 blood samples of 324 sheep with persistent diarrhea from a grassland pasturing area in Xilingol League, Inner Mongolia Municipality, China. Homology analysis indicated that CS13T belongs to the genus Carnobacterium and the closest relatives are Carnobacterium antarticum sp. CP1 (100%), Carnobacterium mobile DSM 4848 (97%) and Carnobacterium funditum DSM 5970 (96%). Similar to those of the C. CP1, the short rod-shaped cells of CS13T are 0.4-0.8 μm wide and 1.0-1.5 μm long; exist singly, paired or catenoid; are gram positive, non-spore forming, and facultatively anaerobic; and produce hemolysin. CS13T cannot produce gas or H2S but can ferment sucrose, galactose, salicin, and esculin to produce acid. However, in contrast to C. CP1, CS13T can produce acid from cellobiose and maltose and is weakly positive for D-mannose fermentation; the growth temperatures range from 20-37°C, the pH range is 5.0-9.0, and the G+C content is 37.84% (4-36°C, pH 6.0-9.5, and 38.1% for C. CP1). Furthermore, based on gene annotation analysis, we found that CS13T has 31 more specific genes than C. CP1 (133 to 102) and that the nonredundant protein similarity to C. CP1 is only 84.2%. Based on the physiological-biochemical and genetic analysis results, the organisms isolated from the Mongolian Plateau and sandy soil in Antarctica belong to the same novel species of the genus Carnobacterium. However, the relationship between CS13T and animal disease needs further study.
Keywords
Environmental Microorganism, Carnobacterium, Diarrhea, Sheep, Mongolian Plateau.
Abstract
Herein, we report a novel Carnobacterium-like organism, CS13T, isolated from Mongolian Plateau is 100% related to the reported Antarctic environmental microorganism Carnobacterium antarticum sp. CP1. The CS13T was isolated from 78 blood samples of 324 sheep with persistent diarrhea from a grassland pasturing area in Xilingol League, Inner Mongolia Municipality, China. Homology analysis indicated that CS13T belongs to the genus Carnobacterium and the closest relatives are Carnobacterium antarticum sp. CP1 (100%), Carnobacterium mobile DSM 4848 (97%) and Carnobacterium funditum DSM 5970 (96%). Similar to those of the C. CP1, the short rod-shaped cells of CS13T are 0.4-0.8 μm wide and 1.0-1.5 μm long; exist singly, paired or catenoid; are gram positive, non-spore forming, and facultatively anaerobic; and produce hemolysin. CS13T cannot produce gas or H2S but can ferment sucrose, galactose, salicin, and esculin to produce acid. However, in contrast to C. CP1, CS13T can produce acid from cellobiose and maltose and is weakly positive for D-mannose fermentation; the growth temperatures range from 20-37°C, the pH range is 5.0-9.0, and the G+C content is 37.84% (4-36°C, pH 6.0-9.5, and 38.1% for C. CP1). Furthermore, based on gene annotation analysis, we found that CS13T has 31 more specific genes than C. CP1 (133 to 102) and that the nonredundant protein similarity to C. CP1 is only 84.2%. Based on the physiological-biochemical and genetic analysis results, the organisms isolated from the Mongolian Plateau and sandy soil in Antarctica belong to the same novel species of the genus Carnobacterium. However, the relationship between CS13T and animal disease needs further study.
Keywords
Environmental Microorganism, Carnobacterium, Diarrhea, Sheep, Mongolian Plateau.
Introduction
Carnobacteria are ubiquitous lactic acid bacteria (LAB), tolerant to freezing/thawing and high pressure and able to grow at low temperatures [1]. The genus belongs to the family Carnobacteriaceae of the phylum Firmicutes, class Bacilli, order Lactobacillales, as described in Bergey’s Manual of Systematic Bacteriology [2], and includes motile, psychrotolerant, short rod-shaped, gram-positive, facultatively anaerobic, heterofermentative lactic acid bacteria that can produce L-lactic acid from mostly fermented D-glucose [3]. At the time of writing, 12 recognized species had been correctly named and collected in the List of Prokaryotic names with Standing in Nomenclature (LPSN) collection (http://www.bacterio.net).
The species C. divergens, C. gallinarum and C. mobile are frequently encountered in the environment and in foods. C. antarcticum, C. alterfunditum, C. funditum and C. iners were isolated from sandy soil, anoxic lake water and the littoral zone of Antarctica [2,4,5]. C. inhibens and C. maltaromaticum were found in Atlantic salmon and infected Lake Whitefish, respectively [6,7]. Additionally, C. pleistocenium was isolated from permafrost of the Fox Tunnel in Alaska [8], C. viridians was isolated from vacuum-packed bologna sausage [9], and C. jeotgali was isolated from a Korean traditional fermented food [10]. Although a large number of research studies have reported isolation of these bacteria from various regions and environments, many species have not yet been allocated to known species, such as the Carnobacterium-like organisms isolated from the larval midgut of a moth species [11], spent mushroom compost [12] and watershed polluted with horse manure [13].
In this study, we isolated a novel Carnobacterium-like organism, designated CS13T, from the blood of sheep with persistent diarrhea in the Mongolian Plateau in China. To further clarify the diversity of this novel isolated strain and Carnobacterium antarticum sp. CP1, this paper discussed the similarities and differences through culture characteristics, phenotypic characterization, and physiological-biochemical and phylogenetic characteristics.
Materials and Methods
Ethics statement
This study was approved by the Animal Ethics Committee of Chongqing Academy of Animal Sciences. The protocol of blood sample collection was established according to A Good Practice Guide to the Administration of Substances and Removal of Blood, Including Routes and Volumes [14].
Collection of blood samples and isolation of strains
Grazing sheep with persistent diarrhea were found at Zhenglan Banner, Xilinguole League, Inner Mongolia Municipality, China (N42°42′09″, E116°13′19″), in 2019. Cervical vein blood samples of sheep were collected by a sterile syringe, and the samples were stored in anticoagulative tubes at 4°C. To culture and separate the pathogens from blood samples, brain heart infusion broth liquid (BHI) medium and Columbia blood agar (CBA, supplemented with 5% (v/v) defibrinated sheep blood) medium were prepared as previously described [15]. Aliquots of 100 μL of the blood samples were streak-inoculated on CBA medium at 4ºC, 20ºC, 25ºC, 30ºC or 37ºC in the presence or absence of oxygen. Colonies were observed after 72 h, and the clearest colonies were subcultured into BHI medium and then cultured for 48 h. Recovered pure cultures were preserved at -80°C in BHI broth supplemented with 20% glycerol.
Physiology and biochemistry observation
To define the optimal culture conditions, the isolated strain was inoculated in BHI medium with extra NaCl concentrations of 1.0-10.0% (at intervals of 1.0%, w/v) at pH 5.0-10.0 (at intervals of 1.0) at growth temperature for 72 h separately. CBA was used as a growth and hemolysin examination medium to culture the isolated strain. Gram staining was conducted with a gram staining kit (Solarbio) and observed by optical microscopy (Nikon). The morphology, size and flagellum ultrastructure of the isolated strain were observed by a JEOL JEM-1200EX electron microscope after uranyl acetate and citromalic acid lead double staining. The biochemical properties, including glycolysis reaction, indole production, hydrogen sulfide production, methyl red test, pyruvate utilization, nitrate reduction and acid production, were determined using a Micro-Biochemical Identification Tube (Hopebio).
Homology and phylogenetic analyses
The genomic DNA of the isolated strain was extracted using a Bacterial DNA Kit (TIANGEN) and then submitted to Sangon Biotech (Shanghai) for sequencing. Homologous sequences were compared with NT (NCBI nucleotide sequences database), NR (NCBI nonredundant protein sequences database) and Swiss-Prot (manually annotated and reviewed protein sequences database). Phylogenetic analysis was performed via maximum-likelihood, maximumparsimony and neighbor-joining algorithms in MEGA version 7.0 [16]. Additionally, comparisons of the core genes, dispensable genes and specific genes were also used in phylogenetic analyses.
Results
Isolation and identification
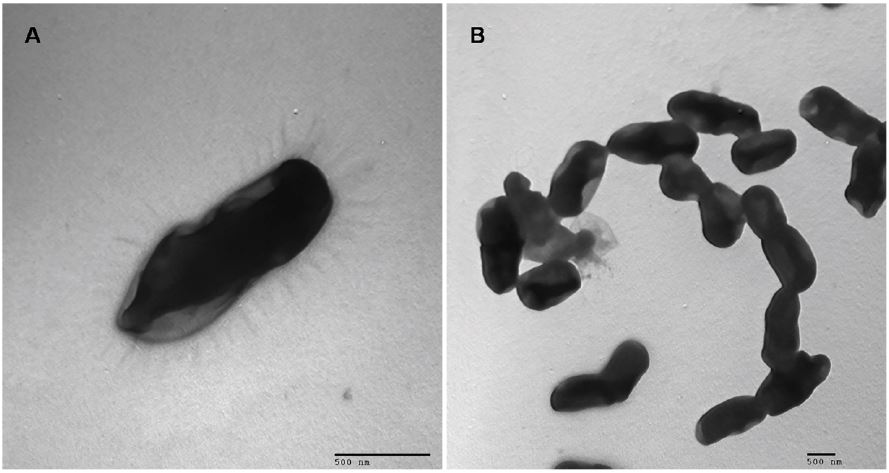
Earlier isolates on CBA medium incubated with oxygen at 20-37°C (optimum, 30°C) for 72 h presented bacterial colonies 1-2 mm in diameter that were white-gray and opaque, with neat edges; had a smooth convex elevation and were surrounded by a tiny hemolysis halo. Furthermore, the growth of bacteria was observed at pH 5.0-9.0 (optimum pH=8.0) and in the presence of 0-5% (w/v) NaCl (optimum, 1%) when the isolated strain was inoculated in BHI media with different pH and salinity values. Electron microscopy demonstrated that the cells of the isolated strain were slightly curved short rods approximately 0.4-0.8 ìm wide and 1.0-1.5 ìm long, with flagella occurring singly or in pairs or short chains (Figure 1).
Physiology and biochemistry
The isolate CS13T has biochemical and physiological characteristics similar to those of the Antarcticaisolated strains Carnobacterium antarticum sp. CP1 and Carnobacterium funditum DSM 5970 and the frozen meatisolated strain Carnobacterium mobile DSM 4848. The four strains exhibit short rod shapes, positive Gram staining, motility, facultatively anaerobic growth, growth at low temperatures, negative oxidase and catalase activities, and no H2S production. In contrast, C. DSM 4848 is the only strain that produces gas, and except C. DSM 5790, all of them utilized esculin, D-glucose, D-mannose, N-acetyl-glucosamine, salicin and sucrose to produce acid. The physiological similarity between the isolates and their closest relatives among the genus are presented in Table 1.
Figure 1:: Electron Microscope Observation. Negative stain electron microscopy of single (A) and pairs (B) of isolated strains.
Homology analysis
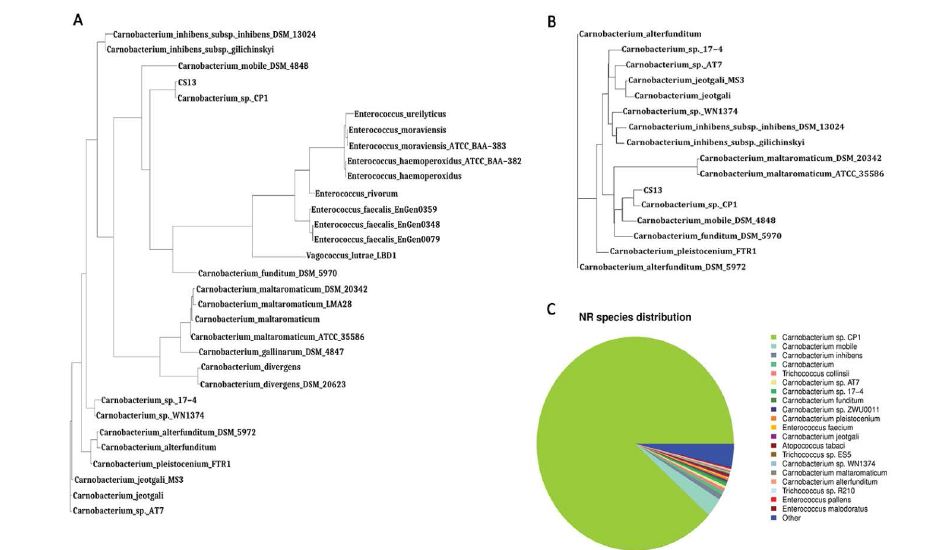
The 16S rRNA gene of CS13T was sequenced by Sangon Biotech (Shanghai). 16S rRNA sequence alignment (NCBI blastn) showed that CS13T shares 100% identity with Carnobacterium antarticum sp. CP1, 97% with Carnobacterium mobile DSM 4848 and 96% with Carnobacterium funditum DSM 5970. The 16S rRNA gene and pangenome phylogenetic trees (Figure 2A,2B) also demonstrated that CS13T and CP1 have the closest relationship. Therefore, we infer that the organism isolated from the blood of sheep on the Mongolian Plateau belongs to the genus Carnobacterium and that CS13T is on the same branch as C. CP1.
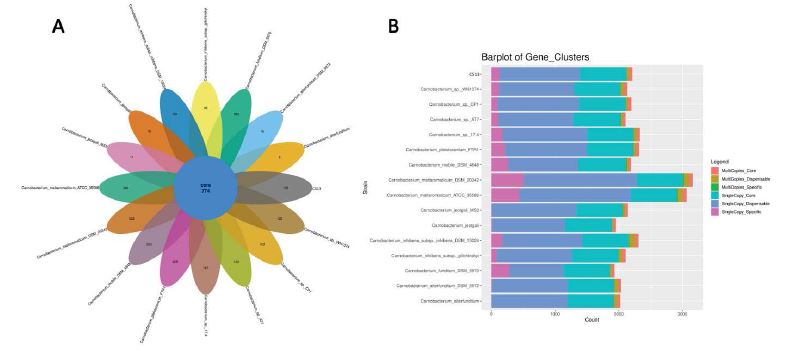
In contrast, the NR species distribution results indicated that CS13T had only 84.2% of protein coding genes that matched those of C. CP1 (Figure 2C). Orthologous cluster analysis revealed that CS13T has 133 specific genes, in comparison with C. CP1 (102 specific genes) and other related species (Figure 3A,3B), indicating that strain CS13T could be classified as a novel species of the genus Carnobacterium.
The protein sequence alignment of CS13T and C. CP1 was carried out by the Circos package (Figure 4). The results illustrated that although the protein sequences of CS13T and C. CP1 are also highly homologous, there are many specific fragments of CS13T, such as NODE_88, NODE_101, NODE_113, NODE_115 and NODE_145, which further confirms CS13T as a novel species.
Discussion
Carnobacterium is found mostly in the intestines of animals. As intestinal probiotics, certain Carnobacterium can also effectively inhibit pathogens and spoilage microorganisms, so they are widely used as a food additive. Among 12 species of Carnobacterium, C. divergens and C. maltaromaticum are frequently isolated from food, particularly in vacuumpackaged (VP) meat and meat products, which also show the ability to inhibit pathogenic and spoilage microorganisms in diverse food matrices. Other scholars reported that C. maltaromaticum B26 and C. divergens B33, isolated from the intestine of healthy rainbow trout, are beneficial for enhancing cellular and humoral immune responses, and they were selected as potential probiotics with effectiveness. In this study, the Carnobacterium antarticum sp. CP1-like organism, CS13T, has been detected by PCR in the blood and in the intestinal contents of some sheep with diarrhea; however, the microorganism was sheep only isolated from the blood. This phenomenon is similar to animal diarrhea caused by conditional pathogens such as Proteus mirabilis, which can cause bacteremia through gastroenteritis [17]. However, the results of the physiological, biochemical and genetic analyses indicate that the microorganisms isolated from the Mongolian Plateau and Antarctica belong to the same novel species of the genus Carnobacterium. We suggest that this species is probably distributed globally and adapts to varying areas and surroundings. Carnobacterium species that have been isolated from the environment and are generally not characterized as pathogens, although they are known to cause disease in fish [18]. Only three reports describing Carnobacterium species from human infection have been found in the literature [19]. Here we report what we believe to be the first isolation and characterization of Carnobacterium species from bacteremia in sheep. Whether this novel species is related to animal disease or can be used in the food industry or other fields remain to be further studied.
Funding Information
This work was funded by the Science and Technology Program of Chongqing Academy of Animal Sciences (19538), Chongqing, China.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Table 1: Differential Physiological Characteristics Distinguishing Strain CS13T from the Closest Members of the Genus Carnobacterium. +, -, W and ND Represent Positive, Negative, Weakly Positive and ‘No Data’, Respectively.
| Characteristic | CS13T | C. CP1* | C. DSM 4848T† | C. DSM 5970T‡ |
|---|---|---|---|---|
| Isolation source | Sheep blood in Mongolian Plateau | Sandy soil in Antarctica | Frozen chicken meat | Lake water in Antarctica |
| Rods | + | + | + | + |
| Growth temperatures (°C) (optimum) | 20-37 (30) | 4-36 (28-32) | 0-35 | 4-20 (4) |
| pH range (optimum) | 5.0-9.0 (8.0) | 6.0-9.5 (8.0-8.5) | ND | ND |
| NaCl tolerance range (w/v) | 0-5.0 (1.0) | 0-5.0 (1.0) | ND | 1-2 |
| Facultatively anaerobic | + | + | + | + |
| Hemolysis | + | + | ND | ND |
| Gram staining | + | + | + | + |
| Motility | + | + | + | + |
| Catalase | - | - | ND | - |
| Oxidase | - | - | ND | ND |
| Produce gas | - | - | + | ND |
| Produce H2S | - | - | - | ND |
| Acid from | ||||
| Cellobiose | + | - | + | - |
| D-Glucose | + | + | + | - |
| D-Mannose | W | + | + | - |
| Maltose | + | - | + | - |
| N-acetyl-glucosamine | + | + | + | - |
| Sucrose | + | + | + | - |
| Salicin | W | W | + | - |
| D-galactose | + | - | + | - |
| Arabitol | - | - | - | - |
| Glycogen | - | - | - | - |
| Starch | - | - | ND | - |
| Xylitol | - | - | - | - |
| Glycerol | W | W | ND | - |
| Esculin | W | W | ND | + |
| DNA G+C content (%) | 37.84 | 38.1 | 35.5-37.2 | 34 |
| * Data for C. antarticum were taken from Zhu et al. [2]. † Data for C. mobile were taken from Collins et al. [15]. ‡Data for C. iners were taken from Snauwaert et al. [5]. | ||||
Figure 2: Homology Analysis. Evolutionary relationships based on the 16S rRNA Gene (A) and pangenome (B) of strain CS13T and related species. (C) The nonredundant protein gene annotation of strain CS13T.
Figure 3:Pan Genomic Analysis. (A). Quantity of core genes (center) and specific genes (petal) of strain CS13T and related species. (B). Gene clusters of strain CS13T and related species.
Leisner JJ, Laursen BG, Prévost H, Drider D, Dalgaard P (2007) Carnobacterium: positive and negative effects in the environment and in foods. FEMS Microbiol Rev 31: 592-613. [ Ref ]
Zhu S, Lin D, Xiong S, Wang X, Xue Z, et al. (2018) Carnobacterium antarcticum sp. nov., a psychrotolerant, alkaliphilic bacterium isolated from sandy soil in Antarctica. Int J Syst Evol Microbiol 68: 1672-1677. [ Ref ]
Nicholson WL, Zhalnina K, de Oliveira RR, Triplett EW (2015) Proposal to rename Carnobacterium inhibens as Carnobacterium inhibens subsp. inhibens subsp. nov. and description of Carnobacterium inhibens subsp. gilichinskyi subsp. nov., a psychrotolerant bacterium isolated from Siberian permafrost. Int J Syst Evol Microbiol 65: 556-561. [ Ref ]
Franzmann PD, Hpfl P, Weiss N, Tindall BJ (1991) Psychrotrophic, lactic acid-producing bacteria from anoxic waters in Ace Lake, Antarctica; Carnobacterium funditum sp. nov. and Carnobacterium alterfunditum sp. nov. Arch Microbiol 156: 255-262. [ Ref ]
Snauwaert I, Hoste B, De Bruyne K, Peeters K, De Vuyst L, et al. (2013) Carnobacterium iners sp. nov., a psychrophilic, lactic acid-producing bacterium from the littoral zone of an Antarctic pond. Int J Syst Evol Microbiol 63: 1370-1375. [ Ref ]
Cantas L, Fraser TWK, Fjelldal PG, Mayer I, Sørum H (2011) The culturable intestinal microbiota of triploid and diploid juvenile Atlantic salmon (Salmo salar) - a comparison of composition and drug resistance. BMC Vet Res 7: 71. [ Ref ]
Loch TP, Xu W, Fitzgerald SM, Faisal M (2008) Isolation of a Carnobacterium maltaromaticum-like bacterium from systemically infected lake whitefish (Coregonus clupeaformis). FEMS Microbiol Lett 288: 76-84. [ Ref ]
Pikuta EV, Marsic D, Bej A, Tang J, Krader P, et al. (2005) Carnobacterium pleistocenium sp. nov., a novel psychrotolerant, facultative anaerobe isolated from permafrost of the Fox Tunnel in Alaska. Int J Syst Evol Microbiol 55: 473-478. [ Ref ]
Holley RA (2002) Carnobacterium viridans sp. nov., an alkaliphilic, facultative anaerobe isolated from refrigerated, vacuum-packed bologna sausage. Int J Syst Evol Microbiol 52: 1881-1885. [ Ref ]
Kim MS, Roh SW, Nam YD, Yoon JH, Bae JW (2009) Carnobacterium jeotgali sp. nov., isolated from a Korean traditional fermented food. Int J Syst Evol Microbiol 59: 3168-3171. [ Ref ]
Shannon AL, Attwood G, Hopcroft DH, Christeller JT (2001) Characterization of lactic acid bacteria in the larval midgut of the keratinophagous lepidopteran, Hofmannophila pseudospretella. Lett Appl Microbiol 32: 36-41. [ Ref ]
Ntougias S, Zervakis GI, Kavroulakis N, Ehaliotis C, Ehaliotis C, et al. (2004) Bacterial diversity in spent mushroom compost assessed by amplified rDNA restriction analysis and sequencing of cultivated isolates. Syst Appl Microbiol 27: 746-754. [ Ref ]
Simpson JM, Santo Domingo JW, Reasoner DJ (2004) Assessment of equine fecal contamination: the search for alternative bacterial sourcetracking targets. FEMS Microbiol Ecol 47: 65-75. [ Ref ]
Diehl KH, Hull R, Morton D, Pfister R, Rabemampianina Y, et al. (2001) A good practice guide to the administration of substances and removal of blood, including routes and volumes. J Appl Toxicol 21: 15-23. [ Ref ]
Zhu S, Wang X, Zhang D, Jing X, Zhang N, et al. (2016) Complete genome sequence of hemolysin-containing Carnobacterium sp. strain CP1 isolated from the Antarctic. Genome Announc 4: e00690-16. [ Ref ]
Collins MD, Farrow JAE, Phillips BA, Feresu S, Jones D (2008) Classification of Lactobacillus divergens, Lactobacillus piscicola, and some catalase-negative, asporogenous, rod-shaped bacteria from poultry in a new genus, Carnobacterium. Int J Syst Evol Microbiol 58: 2672-2672. [ Ref ]
Mobley HLT (2019) Proteus mirabilis Overview. Methods Mol Biol 2021: 1-4. [ Ref ]
Roh H, Kim BS, Lee MK, Park CI, Kim DH (2020) Genome-wide comparison of Carnobacterium maltaromaticum derived from diseased fish harbouring important virulence-related genes. J Fish Dis. [ Ref ]
Hoenigl M, Grisold AJ, Valentin T, Leitner E, Zarfel G, et al. (2010) Isolation of Carnobacterium sp. from a human blood culture. J Med Microbiol 59: 493-495. [ Ref ]