Journal Name: Journal of Applied Microbiological Research
Article Type: Research
Received date: 11 August, 2020
Accepted date: 24 August, 2020
Published date: 31 August, 2020
Citation:
Copyright:
Abstract
Background: Yersinia enterocolitica is a human-animal-fishassociated infectious diarrhea pathogen that has caused widespread international attention in recent years. Many strains of Yersinia enterocolitica were identified from different animal species, but there is no information reported Yersinia enterocolitica in Gymnocypris przewalskii. The Gymnocypris przewalskii is a very important species of an aquatic economic animal in the Qinghai Lake. They were listed in the China’s second-class protected animal species. Preliminary research on the distribution of Yersinia enterocolitica and the drug sensitive fauna test on Gymnocypris przewalskii was an urgent solving problem for which maintain the original ecological symbiotic system and restore Gymnocypris przewalskii resource. In order to solve these issues, we performed this research.
Results: The results showed that 5 strains Yersinia enterocolitica were obtained from Gymnocypris przewalskii. The positive ratio was 6.67% (5/75). The average drug-resistant of 5 strain Yersinia enterocolitica was 54.29% (38/70) to 14 kinds of antibiotic. The result of 16S rRNA gene of Yersinia enterocolitica identified showed that one-piece of 1419 bp specific braid was obtained for the isolate. The homologies of nucleotide of 16S rRNA were 91%-95% between 15 strains of Yersinia enterocolitica from GenBank by measuring sequence.
Conclusion: Yersinia enterocolitica can infect Gymnocypris przewalskii. Some strains have drug-resistant effect. The 16S rRNA gene matches 91%-95% with other strains nucleotides download from GenBank and forms a unique branch separated from them.
Keywords
Fish, Yersinia enterocolitica, Isolation; Drug sensitive fauna test.
Abbreviations
PBS: Phosphate Buffer Solution; GLU: Glucose; LAC: Lactose; MAL: Maltose; SUC: Sucrose; RHA: Rhamnose; ARA: Arabinose; XYL: xylose; MAN: Mannitol; SOR: Sorbitol; DUL: Dulcite; SAL: Saligenin; INO: Inositol; ODC: Ornithine decarboxylase culture; TRP: Tryptophan; CIT: Citrate; NIT: Nitrate; URE: Urease; MR: Methyl red test ; IND: Indol; AZI: Azithromycin; GMIO: Gentamicin; CH: Cephalosporin; E: Erythrocin; AM: Amikacin; CAZ: Cftazidime; PIP: Piperacillin; TE: Tetracycline; TM: Tobramycin; CMZ: Cefuroxime; K: Kanamycin; S300: Streptomycin; SXT: Selectrin; C: Chloromycetin
Abstract
Background: Yersinia enterocolitica is a human-animal-fishassociated infectious diarrhea pathogen that has caused widespread international attention in recent years. Many strains of Yersinia enterocolitica were identified from different animal species, but there is no information reported Yersinia enterocolitica in Gymnocypris przewalskii. The Gymnocypris przewalskii is a very important species of an aquatic economic animal in the Qinghai Lake. They were listed in the China’s second-class protected animal species. Preliminary research on the distribution of Yersinia enterocolitica and the drug sensitive fauna test on Gymnocypris przewalskii was an urgent solving problem for which maintain the original ecological symbiotic system and restore Gymnocypris przewalskii resource. In order to solve these issues, we performed this research.
Results: The results showed that 5 strains Yersinia enterocolitica were obtained from Gymnocypris przewalskii. The positive ratio was 6.67% (5/75). The average drug-resistant of 5 strain Yersinia enterocolitica was 54.29% (38/70) to 14 kinds of antibiotic. The result of 16S rRNA gene of Yersinia enterocolitica identified showed that one-piece of 1419 bp specific braid was obtained for the isolate. The homologies of nucleotide of 16S rRNA were 91%-95% between 15 strains of Yersinia enterocolitica from GenBank by measuring sequence.
Conclusion: Yersinia enterocolitica can infect Gymnocypris przewalskii. Some strains have drug-resistant effect. The 16S rRNA gene matches 91%-95% with other strains nucleotides download from GenBank and forms a unique branch separated from them.
Keywords
Fish, Yersinia enterocolitica, Isolation; Drug sensitive fauna test.
Abbreviations
PBS: Phosphate Buffer Solution; GLU: Glucose; LAC: Lactose; MAL: Maltose; SUC: Sucrose; RHA: Rhamnose; ARA: Arabinose; XYL: xylose; MAN: Mannitol; SOR: Sorbitol; DUL: Dulcite; SAL: Saligenin; INO: Inositol; ODC: Ornithine decarboxylase culture; TRP: Tryptophan; CIT: Citrate; NIT: Nitrate; URE: Urease; MR: Methyl red test ; IND: Indol; AZI: Azithromycin; GMIO: Gentamicin; CH: Cephalosporin; E: Erythrocin; AM: Amikacin; CAZ: Cftazidime; PIP: Piperacillin; TE: Tetracycline; TM: Tobramycin; CMZ: Cefuroxime; K: Kanamycin; S300: Streptomycin; SXT: Selectrin; C: Chloromycetin
Introduction
Yersinia enterocolitica is a human-animal-fish-associated infectious diarrhea pathogen that has caused widespread international attention in recent years. Yersiniasis caused by Yersinia enterocolitica is the third most common zoonotic disease followed by Salmonella and Campylobacter in some European countries [1]. It can often cause the respiratory, cardiovascular systems, bones and connective tissues diseases [2,3], making the disease with poor prognosis or death due to sepsis [4]. At present, the bacteria have been isolated from various mammals, oviparous animals, agricultural products and animal products in China and abroad [5-10]. However, no reports of Yersinia enterocolitica from Gymnocypris przewalskii in Qinghai Lake have been seen.
The Gymnocypris przewalskii belongs to the Gymnocypris, Schizothoracinae, Cyprinidae, Cypriniformes and common called ‘Huangyu’. It is the only aquatic economic animal in Qinghai lake and its water system. At the end of the last century, due to the shrinking water area of the Qinghai Lake, the deterioration of the water environment, the frequent occurrence of various diseases and the overfishing of spawning and spawning, the phenomenon of spawning and spawning of broods’ tock was repeatedly banned. In addition, the hatching rate of fertilized eggs of broods’ tock in the natural state was less than 5% and their growth was very slow (Gains about 0.15 kg every 10 years), leading to a sharp decline in the population base to about 2000 tons. With the increasing intensity of lake closure and artificial seedling release in Qinghai Lake in recent years, although the population base has reached 88,000 tons, it is still less than one third of the 1960’s. At present, researches on Gymnocypris przewalskii in Qinghai Lake are mostly focused on biology and parasitic diseases, while research on pathogenic microorganisms is almost blank.
Methods
Samples collection
75 pancreas of Qinghai Lake naked carp that had died one after another in a fishery of a naked carp rescue center in Qinghai Province were aseptically collected and numbered, and placed in a refrigerator at 4ºC for use.
Control strains
Escherichia coli (ATCC25922) and Salmonella typhimurium (ATCC14028) were donated by the Department of Preventive Veterinary Medicine, China Agricultural University.
Growth medium and molecular reagents
The Improved Phosphate Buffer Solution (PBS), modified Y and CIN-1 plates, and modified Kirschner’s disaccharide iron bevel (KIA) were made in our laboratory. Glucose and other 24 biochemical media were purchased from Beijing Luqiao Technology Co., Ltd. (batch number: 20180611). Bacterial DNA extraction kit, gel recovery kit, Taq DNA polymerase and pMD18-T vector were purchased from TaKaRa.
Primers
The primers sequences are P1 5’CGCGGATCCATTGAACGCTGGCGGCAG3’ and P2 5’GGGGTACCCCTACGGTTACCTTGTTACGACTTC3’. They were synthesized by Shanghai Biotech Biotechnology Service Co., Ltd.
Drug sensitive paper
14 kinds of drug sensitive paper such as chloramphenicol were purchased from Hangzhou Microbial Reagent Co., Ltd. (Lot No.: 20180290).
Isolation and identification of Yersinia enterocolitica from naked carp of Qinghai Lake.
Sample enrichment: Cut a few samples aseptically and inoculate them in PSB, and incubate them at 26ºC for 48 ± 2h.
Sample alkali treatment: Take 1 mL of the bacterial growth solution and add it to 9 mL of 0.4% KOH solution (containing 0.5% NaCl, prepared before use), which is warmed to 26ºC, and mix for 30 s.
Isolation and purification of bacteria: Accordion to the method recommended by Aulisio [11]. Alkali-treated bacteria-enriching solution was streaked on CIN-1 and modified Y plates, and cultured at 26ºC for 48 ± 2 h. After that, the suspect colony smears were picked, and Gram staining microscopy was performed, and several purification cultures were performed.
Screening of bacteria
Five suspicious colonies on CIN-1 and modified Y plate were picked in turn and tested on KIA oblique urea and semi-solid medium, and the results were observed.
Smear, Gram stain and microscopy
Bacteria that were positive at 26ºC and negative at 37ºC in the secondary rescreening test were picked for smear and Gram staining microscopy.
Biochemical identification
The suspected strains screened by the primary and secondary screening tests were aseptically inoculated into 27 different tubes, such as glucose. Then cultured those at 26ºC for 48 ± 2h, and the results were observed.
Drug sensitive test
According to the method by Meyer [12].The purified strains were individually inoculated into a modified Y slant, cultured at 26ºC for 24 hours, and then washed under sterile conditions with sterile physiological saline, and were turbid with a Macquarie tube. A bacterial suspension at a concentration of 9 × 108 live bacteria / mL was selected, and the bacterial solution was dipped in a sterile cotton swab to uniformly coat the MH (A) plate 3 times. Then, 16 kinds of drug-sensitive papers were respectively put on in order, 3 pieces were put on each plate, and the results were determined by incubating at 37ºC for 18-24h.
Cloning of 16S rRNA gene
Template DNA Extraction: The operation steps are performed according to the instruction manual of MiniBEST Bacterial Genomic DNA Extraction Kit.
PCR reaction system and conditions: According to and modified the method by Neubauer [13].PCR reaction system (50 μL): Premix Taq 25 μL, P1 and P2 primers 1 μL each, DNA template 1 μL, ddH2O 22 μL; PCR reaction conditions: 94ºC pre-denaturation for 5 min, 94ºC denaturation for 30 s, 55ºC 30 s, 72ºC for 30 s, a total of 35 cycles, 72ºC extended 10 minutes.
16S rRNA Gene Cloning and Sequence Analysis:The PCR amplified product recovered from the gel was cloned, PCR identified and sequenced 3 times. The 16S rRNA sequence of Yersinia enterocolitica from naked carp of Qinghai Lake was performed on GenBank and 15 representative strains of China and abroad Homology analysis.
Results
Routine isolation and identification of Yersinia enterocolitica from naked carp of Qinghai Lake
Colony morphology and culture characteristics: Red bull-eye colonies appeared on CIN-1. Colorless and transparent, non-sticky colonies appeared on modified Y. Gram staining microscopic examination showed short rodshaped Gram-negative bacilli or cocci, mostly scattered individually, sometimes arranged in short chains or piles, without spores and capsules.
Screening of bacteria
After preliminary screening tests, 20 strains of KIA with yellow and non-gas-producing strains were obtained on the slope and bottom of KIA. Fifteen urea-positive strains were obtained by rescreening. Two rescreening yielded 5 strains of 26ºC cultured with motility, and 37ºC cultured non-motility strains, which were classified as Y1301, Y1302, Y1303, Y1304, Y1305.
Biochemical identification
See Table 1 for details.
Drug sensitive test
The results are as follows (Table 2)
16S rRNA gene clone
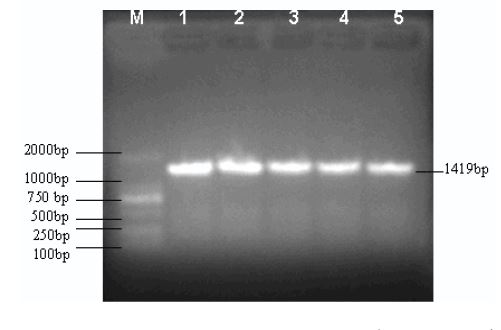
PCR amplification of 16S rRNA gene of Yersinia enterocolitica from naked carp of Qinghai Lake (Figure 1).
PCR specificity test of wild isolates and control bacteria
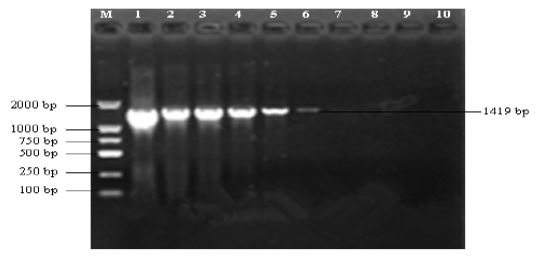
The PCR specificity test of 16s rRNA gene was performed on Y1301 and Y1302 strains with P1 and P2 primers, respectively. The target fragment of 1419 bp in size consistent with the expected results was obtained (Figure 2).
Sensitivity test of isolated strain
A single clone of Y1301 was aseptically inoculated into LB Amp + liquid medium, cultured at 26ºC for 12 hours, and genomic DNA was extracted. The extracted DNA was diluted 10-fold to 10-10 in turn, and P1 and P2 primer pairs were applied respectively. The PCR amplification of 16S rRNA gene was performed, and the obtained PCR product was subjected to electrophoresis, and the results showed that a dilution of 10-6 was detectable (Figure 3).
Homologous analysis of sequence of Y.e 16S rRNA gene from naked carp of Qinghai Lake
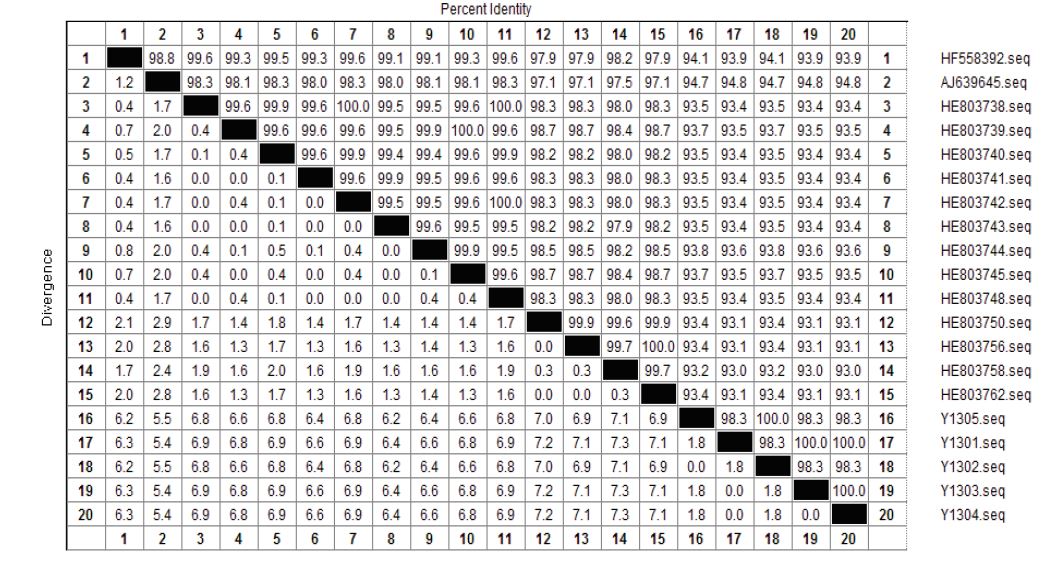
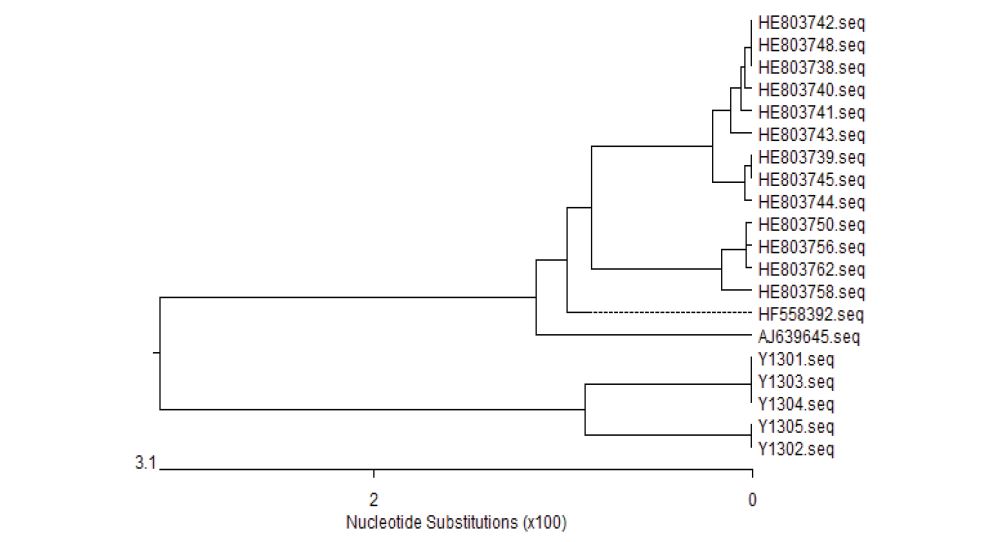
The nucleotide sequence comparison showed that the nucleotide homology of Y1301, Y1303 and Y1304 was 100%; the homology of Y1302 and Y1305 was 100%, and there were no mutation sites. AJ639645, HE803738, HE803739, HE803740, HE803741, HE803742, HE803743, HE803744, HE803745, HE803748, HE803750, HE803756, HE803758, HE803762, HE803792 are in sites of 28th, 72th, 75th, 81th, 84th, 101th, 112nd, 154, 299 th, 325 th, 346 th, 347 th, 351st , 352nd , 359 th, 361 st, 362 nd, 366 th, 367 th, 481 st, 485 th, 491 st, 507 th, 511 st, 515 th, 529 th, 540th, 702 nd, 729 th, 739th, 806 th, 816 th, 867 th, 892 nd, 893 th, 897 th, 912 nd, 913 th, 928 th, 929 th, 992-997 th, 998 th -1006 th, 118 th, 1010 th -1012 nd, 1016 th -1017 th 1020 th-1025th, 1041st -1042 nd, 1046 th 1050 th - 1055 th, 1057 th -1067 th, 1070 th -1073 th, 1076 th -1077 th, 1078 th -1079 th, and 1081st -1082nd have changed. Y1301 had base deletions at 915 and 976, and Y1302 had base deletions at 816 and 867, respectively (Figure 4 and Figure 5).
Figure 1:PCR Amplification of gene of 16S rRNA (Y1301, Y1302 ) by use of P1, P2 Primer. M:DL 2000 Marker;1:Y1301;2:Y1302 ;3:Y1303;4:Y1304;5:Y1305.
Figure 2:Specificity of single PCR of Y1301, Y1302. M:DL 2000 Marker;1:Y1301;2:Y1302;3:Escherichia coli(ATCC25922) ;4:Salmonella typhimurium (ATCC14028).
Figure 3:Sensitivity of single PCR of Y1301. M:DL 2000 Marker;1 :10-1;2:10-2;3:10-3;4:10-4;5:10-5;6:10-6;7:10-7;8:10-8 ;9:10-9;10:10-10
Table 1: The result of biochemical characteristics of Yersinia enterocolitica.
| Items | Y1301 | Y1302 | Y1303 | Y1304 | Y1305 | |
|---|---|---|---|---|---|---|
| GLU | + | + | + | + | + | |
| LAC | - | - | - | - | - | |
| MAL | - | - | - | - | - | |
| SUC | + | + | + | + | + | |
| Trehalose | + | - | + | + | - | |
| RHA | - | - | - | - | - | |
| Raffinose | - | - | - | - | - | |
| ARA | + | - | + | + | - | |
| XYL | - | - | - | - | - | |
| MAN | + | + | + | + | + | |
| SOR | + | + | + | + | + | |
| DUL | - | - | - | - | - | |
| SAL | - | - | - | - | - | |
| INO | + | + | + | + | + | |
| ODC | + | + | + | + | + | |
| TRP | - | - | - | - | - | |
| CIT | - | - | - | - | - | |
| NIT | + | - | + | + | - | |
| URE | + | + | + | + | + | |
| Semi-solid | 26°C | + | + | + | + | + |
| 37°C | - | - | - | - | - | |
| GEL | + | + | + | + | + | |
| MR | - | - | - | - | - | |
| V-P | 26°C | + | + | + | + | + |
| 37°C | - | - | - | - | - | |
| H2S | - | - | - | - | - | |
| IND | - | - | - | - | - | |
| Note: "+" means positive, "-" means negative | ||||||
Table 2: Sensitive Test of 14 kinds of Medicine for 5 strains Yersinia enterocolitis.
| Antibiotics | Y1301 | Y1302 | Y1303 | Y1304 | Y1305 | Resistance | intermediary | Sensitivity |
|---|---|---|---|---|---|---|---|---|
| AZI | S | R | R | S | R | 60% | 0.00% | 40% |
| GMIO | S | S | S | S | I | 0.00% | 20% | 80% |
| CH | R | R | R | R | R | 100% | 0.00% | 0.00% |
| E | R | R | S | I | R | 60% | 20% | 20% |
| AM | S | R | S | R | I | 40% | 20% | 40% |
| CAZ | R | R | I | R | S | 60% | 20% | 20% |
| PIP | R | S | R | S | I | 40% | 20% | 40% |
| TE | I | I | S | R | I | 20% | 60% | 20% |
| TM | S | S | S | R | S | 20% | 0.0% | 80% |
| CMZ | R | R | R | R | S | 80% | 0.00% | 20% |
| K | R | R | R | S | I | 60% | 20% | 20% |
| S300 | R | R | R | I | R | 80% | 20% | 0.00% |
| SXT | R | R | R | R | I | 80% | 20% | 0.00% |
| C | R | S | R | R | S | 60% | 0.00% | 40% |
Discussion
The results of the drug susceptibility test showed that the resistance rate of five strains to 14 types of drug susceptibility tests exceeded 50.00%. Y1301 is resistant to 9 drugs: CH, E, CAZ, K, AMP, PIP, CMZ, K, P, S300, SXT, and C. The drug resistance rate is 56.25% (9/16). Y1302 is resistant to 11 drugs: AZI, CH, E, AN, CAI, AMP, CMZ, P, K, P, E, S300, and SXT, with a drug resistance rate of 68.75% (11/16). Y1303 has resistance to 10 drugs, AZI, CH, PIP, AMP, K, P, S300, SXT, and C, with a drug resistance rate of 62.50% (10/16). Y1304 has resistance to 9 drugs: CH, AN, CAZ, TE, TM, CMZ, P, SXT, and C. The drug resistance rate is 56.25% (9/16). Y1305 is resistant to five drugs, AZI, CH, E, AMP, P, and S300, with a resistance rate of 31.25% (5/16). The results of the drug sensitivity test showed that the isolate was highly sensitive to two drugs, gentamicin and tobramycin, but resistant to penicillin. Therefore, these high-sensitivity drugs can be selected for prevention and control in production. This result is different from the drug sensitivity characteristics of Yersinia isolated from spot fork tails infected by Y. enterocolitica The possible reason for the difference in drug sensitivity characteristics is that the strains in different regions and different water environments are exposed to the effects of different drug environmental effects, resulting in differences in drug resistance variations [14-18].
Figure 4:Analysis of genetic evolution of Yersinia enterocolitica 16S rRNA gene from naked carp in Qinghai Lake.
Figure 5:Phylogenetic Tree based on nucleotide sequence of 16S rRNA gene.
Because biochemical characteristics of Y. enterocolitica are more complex, accurate biochemical test results are essential to identify Y. enterocolitica. The isolates tested in this test were sucrose-positive, raffinose-negative, VP (26ºC) positive, and VP (37ºC) negative, all of which were in line with typical Y. enterocolitica characteristics [19-21]. In addition, the biochemical characteristics of other bacteria belonging to the genus Enterobacteriaceae were used. The biochemical characteristics of indole-positive and fermented sucrose can be distinguished from Yersinia pseudotuberculosis. At the same time, according to the biochemical characteristics of the isolates and the typical culture characteristics on CIN- 1 and modified Y medium, it can be clearly distinguished from the genera Proteus, Salmonella and Shigella. Finally, according to the Berger’s Bacteria Identification Manual [22] and Miaoying Cai ‘s “Enterobacteriaceae Retrieval System” [23], the isolate was determined to be Y. enterocolitica. Because this bacterium is a psychrophilic pathogen, it brings great difficulty to the isolation and identification. According to the National Standard for “Yersinia enterocolitica Test” (GB4789.8-2008) issued by the Ministry of Health of the People’s Republic of China, by using the alkali treatment method in combination with CIN-1 to select media and improve Y, a better Separate selection effects was succeeded. The detection medium for naked carp’s solid organ Yersinia enterocolitica shortened the inspection time by 3d, saved about 40% of raw materials, and improved the detection rate. On the one hand, it demonstrates the superiority of this method, and on the other hand, it also suggests the carrier rate in local naked carps. Five Yersinia enterocolitica were isolated from the organs and gills of 75 naked carps collected, and the positive rate was 6.67% (5/75).
Yersinia enterocolitica, combined with Yersinia pseudotuberculosis and Yersinia pestis has been shown to infect human and cause disease clearly. The characterization of other eight Yersinia species, usually referred as “Yersinia enterocolitica-like” strains are limited. Up to date, however more and more information inferred that these species have novel virulence mechanisms, and some of them are associated with human disease [24-27]. Identification of these species is a difficult task. The traditional identification of bacteria is based on the phenotypic characteristics, which are not accurate as genotypic methods. The comparison of the 16S rRNA sequence has emerged as a most preferred genotypic method for the identification of bacteria in their genus level. A 16S rRNA gene-based PCR assay for identifying and separating Y. enterocolitica isolates recently has been developed [13].Here we studying the isolates for PCR amplification of 16S rRNA gene of Yersinia enterocolitica from naked carp of Qinghai Lake, PCR specificity test, sensitivity test and homologous analysis of isolate sequence , showed more positive identification results that have shown in specificity, sensitivity and homology of 16S rRNA for diagnosis of Yersinia enterocolitica strains isolated from the lake [28].
Conclusion
In this study, 5 isolated strains of Yersinia enterocolitica were identified via traditional biochemical, Pathogenicity identification and drug resistance testing. The result is that they were identified as Yersinia enterocolitica with severe drug resistance. At the same time, a phylogenetic tree was constructed based on the 16S rRNA gene of Yersinia enterocolitica, and the results showed that Yersinia enterocolitica isolated from Qinghai Lake naked carp located in the Qinghai-Tibet Plateau was an independently evolved cluster. Compared to other strains isolated from non-Tibet Plateau, they show clear regional differences. By routine identification and specific, genetic identification of Yersinia enterocolitica from naked carp of Qinghai Lake and detection of drug-susceptible flora, the interspecies distribution and drug distribution of Yersinia from naked carp of Qinghai Lake can be found out. Sensitive fauna lays a theoretical foundation for future research on key control points for preventing the disease. The theoretical and practical significance of protecting local special animal resources, exploring ways and capabilities to adapt to the environment, and laying a research foundation for comprehensively protecting the biodiversity of the Qinghai-Tibet Plateau.
Declaration
Ethics approval and consent to participate
No specific permits were required for this study.
Availability of data and materials
All data generated or analyzed during this study and supporting the conclusions of this article are included within the article.
Competing Interests
The authors declare that they have no competing interests.
Authors’ Contribution
ZH, CQ and ZJ conceived and designed the experimental concept. FY and KF collected the samples and extracted the DNA. TL and GX conducted the lab experiments. ZH and CQ wrote the paper. All authors reviewed the manuscript approved the final manuscript.
Acknowledgment
The authors would like to express their deep appreciation for the National Natural Science Foundation of China, which provided the funding for this research. We also thank all of the people who supported us in the field and lab during this research.
Funding
Our research was supported by the “the National Natural Science Foundation of China” (Grant No.31560695). By this funding support, we systematically studied the interspecies transmission characteristics and drug-susceptibility flora of Yersinia enterocolitica in Qinghai Lake naked carp. The genetic evolution and drug resistance under various natural and climatic conditions have laid the foundation for the biological characteristics of Yersinia enterocolitica in the Qinghai-Tibet Plateau, the distribution characteristics of virulence genes, the molecular pathogenic mechanism of invasion genes and prevention and control research.
Arguthority EFS (2012) The European Union summary report on trends and resources of zoonoses, zoonotic agents and food-brone outbreaks in 2010. EFSA Journal 10: 2597. [ Ref ]
Lupi A (2013) Subacute endocarditis caused by Yersinia enterocolitica: a case report. Scand J Infect Dis 45: 329-333. [ Ref ]
Mert M, Kocabay G, Ozülker T (2011) Liver abscess due to Yersinia bacteremia in a well-controlled type I diabetic patient. Endokrynol Pol 62: 357-360. [ Ref ]
Guinet F, Carniel E, Leclercq A (2011) Transfusion-transmitted Yersinia enterocolitica sepsis. Clin Infect Dis 53: 583-591. [ Ref ]
Ong KL (2012) Changing epidemiology of Yersinia enterocolitica infections: markedly decreased rates in young black children, Foodborne Diseases Active Surveillance Network (FoodNet), 1996-2009. Clin Infect Dis 54(S4): S385-S390. [ Ref ]
Kiskova J (2011) Yersinia species in the dunnock (Prunella modularis) in sub-alpine habitats of the Western Carpathians. Pol J Microbiol 60: 79-83. [ Ref ]
Foti M (2011) Pathogenic microorganisms carried by migratory birds passing through the territory of the island of Ustica, Sicily (Italy). Avian Pathol 40: 405-409. [ Ref ]
Fuchs TM (2011) Shotgun sequencing of Yersinia enterocolitica strain W22703 (biotype 2, serotype O:9): genomic evidence for oscillation between invertebrates and mammals. BMC Genomics 12: 168. [ Ref ]
Xiao Y, Liang J, Gu W (2012) Isolation and result analysis of Yersinia enterocolitis in different animal hosts. Chinese Journal of Zoonoses 28: 418- 420. [ Ref ]
Zhao J (2013) The main biological characteristics of Yersinia enterocolitis in pelagoba tepeloco. Journal of southwest Minzu university (natural science edition) 39: 12-16. [ Ref ]
Aulisio CC, Mehlman IJ, Sanders AC (1980) Alkali method for rapid recovery of Yersinia enterocolitica and Yersinia pseudotuberculosis from foods. Appl Environ Microbiol 39: 135-140. [ Ref ]
Meyer C, Stolle A, Fredriksson-Ahomaa M (2011) Comparison of broth microdilution and disk diffusion test for antimicrobial resistance testing in Yersinia enterocolitica 4/O:3 strains. Microb Drug Resist 17: 479-484. [ Ref ]
Neubauer H (1999) Variations in the 16S rRNA gene sequence of Yersinia enterocolitica isolates influence the specificity of molecular identification systems. Zentralbl Bakteriol 289: 329-337. [ Ref ]
Kanazawa Y, Kuramata T (1976) Drug sensitivity of Yersinia enterocolitica and Yersinia pseudotuberculosis. Jpn J Antibiot 29: 366-376. [ Ref ]
Meng J (2019) Differential regulation of physiological activities by RcsB and OmpR in Yersinia enterocolitica. FEMS Microbiol Lett. [ Ref ]
Martins, BTF (2018) Yersinia enterocolitica in a Brazilian pork production chain: Tracking of contamination routes, virulence and antimicrobial resistance. Int J Food Microbiol 276: 5-9. [ Ref ]
Peng ZX (2018) [Antimicrobial susceptibility and drug-resistance genes of Yersinia spp. of retailed poultry in 4 provinces of China] Zhonghua Yu Fang Yi Xue Za Zh 52: 358-363. [ Ref ]
Wang WQ (2019) [Antimicrobial resistance and molecular epidemiology of foodborne Yersinia enterocolitica in Pudong New District, Shanghai]. Zhonghua Liu Xing Bing Xue Za Zhi 40: 354-359. [ Ref ]
Sedgwick AK, Tilton RC (1971) Biochemical and serological characteristics of a Yersinia enterocolitica isolate. Appl Microbiol 21: 383-384. [ Ref ]
Odyniec M (2020) Bioserotypes, Virulence Markers, and Antimicrobial Susceptibility of Yersinia enterocolitica Strains Isolated from Free-Living Birds. Biomed Res Int 2020: 1-6. [ Ref ]
Motallebi M, Zamani MR, Saffar B (2000) Serological and biochemical characteristics of virulence plasmid of Yersinia enterocolitica isolates from chicken in the Islamic Republic of Iran. East Mediterr Health J 6: 409-415. [ Ref ]
Brenner (2005) Bergey’s Manual of Systematic Bacteriology. Springer US. [ Ref ]
Miaoying C, Xiuzhu D (1989) Enterobacteriaceae Retrieval System. [ Ref ]
Bottone EJ (1997) Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev 10: 257-276. [ Ref ]
Delor I (1990) Nucleotide sequence of yst, the Yersinia enterocolitica gene encoding the heat-stable enterotoxin, and prevalence of the gene among pathogenic and nonpathogenic yersiniae. Infect Immun 58: 2983-2988. [ Ref ]
Sulakvelidze A (2000) Yersiniae other than Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis: the ignored species. Microbes Infect 2: 497-513. [ Ref ]
Sulakvelidze A (1999) Production of enterotoxin by Yersinia bercovieri, a recently identified Yersinia enterocolitica-like species. Infect Immun 67: 968-971. [ Ref ]
Hallanvuo S (2006) Simplified phenotypic scheme evaluated by 16S rRNA sequencing for differentiation between Yersinia enterocolitica and Y. enterocolitica-like species. J Clin Microbiol 44: 1077-1080. [ Ref ]