Journal Name: Journal of Applied Microbiological Research
Article Type: Research Methods
Received date: 20 June, 2022
Accepted date: 18 July, 2022
Published date: 25 July, 2022
Citation: Saphier M, Moshkovich L, Popov S, Shotland Y, Silberstein E et al. (2022) Monovalent Copper Ions Inhibit Enzymatic Systems. J Appl Microb Res. Vol: 5 Issu: 2 (01-12).
Copyright: © 2022 Saphier M et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
The effect of monovalent copper ions on enzymatic systems has hardly been studied to date; this is due to the low stability of monovalent copper ions in aqueous solutions, which led to the assumption that their concentration is negligible in biological systems. However, in an anaerobic atmosphere, and in the presence of a ligand that stabilizes the monovalent copper ions over the divalent copper ions, high and stable concentrations of monovalent copper ions can be reached. Moreover, the cell cytoplasm has a substantial concentration of potential stabilizers that can explain significant concentrations of monovalent copper ions in the cytoplasm. This study demonstrates the effect of monovalent and divalent copper ions on DNA polymerase, ligaseT4 DNA, the restriction enzymes EcoP15I and EcoR I, acid phosphatase, and α and βamylase enzymes. These systems were chosen because they can be monitored under conditions necessary for maintaining a stable concentration of monovalent copper ions, and since they exhibit a wide range of dependency on ATP. Previous studies indicated that ATP interacts with monovalent and divalent copper ions and stabilizing monovalent copper ions over divalent copper ions. The results showed that monovalent copper ions dramatically inhibit DNA polymerase and acid phosphatase, inhibit ligaseT4 DNA and the restriction enzyme EcoP15I, moderately inhibit α and β amylase, and have no effect on the restriction enzyme EcoR I.
From the results presented in this work, it can be concluded that the mechanism is not one of oxidative stress, even though monovalent copper ions generate reactive oxygen species (ROS). Molecular oxygen in the medium, which is supposed to increase the oxidative stress, impairs the inhibitory effect of monovalent and divalent copper ions, and the kinetics of the inhibition is not suitable for the ROS mechanism.
ATP forms a complex with copper ions (di and monovalent ions, where the latter is more stable) in which the metal ion is bound both to the nitrogen base and to the oxygen charged on the phosphate groups, forming an unusually distorted complex. The results of this study indicate that these complexes have the ability to inhibit enzymatic systems that are dependent on ATP.
This finding can provide an explanation for the strong antimicrobial activity of monovalent copper ions, suggesting that rapid and lethal metabolic damage is the main mechanism of monovalent copper ions’ antimicrobial effect.
Keywords
Cu+ ions, Enzyme inhibitor, DNA Polymerase, PCR; Phosphatase, Restriction enzymes, Ligase, Amylase, Antibacterial effect, ATP complexes.
Abstract
The effect of monovalent copper ions on enzymatic systems has hardly been studied to date; this is due to the low stability of monovalent copper ions in aqueous solutions, which led to the assumption that their concentration is negligible in biological systems. However, in an anaerobic atmosphere, and in the presence of a ligand that stabilizes the monovalent copper ions over the divalent copper ions, high and stable concentrations of monovalent copper ions can be reached. Moreover, the cell cytoplasm has a substantial concentration of potential stabilizers that can explain significant concentrations of monovalent copper ions in the cytoplasm. This study demonstrates the effect of monovalent and divalent copper ions on DNA polymerase, ligaseT4 DNA, the restriction enzymes EcoP15I and EcoR I, acid phosphatase, and α and βamylase enzymes. These systems were chosen because they can be monitored under conditions necessary for maintaining a stable concentration of monovalent copper ions, and since they exhibit a wide range of dependency on ATP. Previous studies indicated that ATP interacts with monovalent and divalent copper ions and stabilizing monovalent copper ions over divalent copper ions. The results showed that monovalent copper ions dramatically inhibit DNA polymerase and acid phosphatase, inhibit ligaseT4 DNA and the restriction enzyme EcoP15I, moderately inhibit α and β amylase, and have no effect on the restriction enzyme EcoR I.
From the results presented in this work, it can be concluded that the mechanism is not one of oxidative stress, even though monovalent copper ions generate reactive oxygen species (ROS). Molecular oxygen in the medium, which is supposed to increase the oxidative stress, impairs the inhibitory effect of monovalent and divalent copper ions, and the kinetics of the inhibition is not suitable for the ROS mechanism.
ATP forms a complex with copper ions (di and monovalent ions, where the latter is more stable) in which the metal ion is bound both to the nitrogen base and to the oxygen charged on the phosphate groups, forming an unusually distorted complex. The results of this study indicate that these complexes have the ability to inhibit enzymatic systems that are dependent on ATP.
This finding can provide an explanation for the strong antimicrobial activity of monovalent copper ions, suggesting that rapid and lethal metabolic damage is the main mechanism of monovalent copper ions’ antimicrobial effect.
Keywords
Cu+ ions, Enzyme inhibitor, DNA Polymerase, PCR; Phosphatase, Restriction enzymes, Ligase, Amylase, Antibacterial effect, ATP complexes.
Introduction
Enzymes are responsible for most life-sustaining biochemical processes. Impairment of enzymatic activity causes great damage to living systems. Bacteria, being small single cells are even more susceptible to a decrease in the activity of enzymes. Metal ions in general, and transition metals ions in particular, may have a significant effect on the activity of the various enzymes, and indeed much knowledge has been accumulated about the effect of ions found in biological systems on enzymatic activity. However, the effect of monovalent copper ions on enzymatic systems has hardly been studied to date, due to the low stability of monovalent copper ions in aqueous solutions, leading to an estimation that their concentration is negligible in biological systems. Despite the above, recent studies show that monovalent copper ions (Cu+) have a pronounced effect on prokaryotic cells [1,2].
Copper (Cu) is an essential trace element for all living organisms; in high concentrations, however, it can exert a biocidal effect. Recently, monovalent copper ions (Cu+) have been suggested to be the active factor in the antimicrobial activity of copper [1]. Although the mechanism of Cu+ ions’ antimicrobial effect is not yet fully understood, recently published results show that conditions of acidic pH, an unfavorable carbon source, and elevated temperatures boost the antibacterial action of Cu+ ions. In less than 1 min, 0.4mM of Cu+ ions eliminated a 106/cm3 E.coli bacterial population; microscopic images of E.coli morphology showed mortality of bacteria with almost no lysis [2].
Silver is also known as a biocidal agent, especially in the form of monovalent silver ions (Ag+) [3]. As copper and silver are similar elements (coins elements), and their monovalent ions have similar electronic configurations (d10), it is reasonable to assume that both ions have similar biocidal mechanisms.
Copper ions (Cu2+ and Cu+) are known to form reactive oxygen species (ROS) that can damage biomolecules, including DNA and chromatin [4]. This has been welldemonstrated in vitro with isolated DNA or chromatin, or by exposure of cultured mammalian cells to copper complexes with various agents [5,6]. In vivo, however, according to the literature, copper ions do not catalyze the formation of oxidative DNA damage [7-9].
Even so, in living cells there are mechanisms that control the intracellular concentrations of copper ions [10]. ATP7b and CopA pump excess copper out of the cytosol and into the periplasm [11]. Once in the periplasm, copper ions are subject to two other systems, CueO and CusCFBA that assist CopA in controlling intracellular copper levels. CueO is a multi-copper oxidase that converts Cu+ ions to Cu2+ ions, a less-toxic form [11]. It seems that copper ions, especially in the form of Cu+, are a major threat to bacterial cells.
Copper ions use as a weapon in the antimicrobial arsenal of grazing protozoa and of phagocytic cells of the immune and affects central carbon metabolism in Staphylococcus aureus, which implies intracellular activity [12].
In aqueous solutions, copper in the monovalent state (Cu+(aq), cuprous) is unstable compared to the common oxidation state of the copper ion, the divalent state (Cu2+ (aq), or cupric). Monovalent copper ions in aqueous solutions undergo disproportion reaction to Cu2+(aq) and Cu0 (s), andin aerobic environments react rapidly with molecular oxygen. As a result, in aqueous solutions monovalent copper ions’ concentrations are usually very low. To achieve significant concentrations of monovalent copper ions in aqueous solutions, a ligand, such as acetonitrile, benzoic acid or ATP, has to be added to the solutions [13-16]. These ligands tilt the existing equilibrium between oxidation states toward the formation of two Cu+ ions from one Cu2+ ion and metallic copper. In addition, the molecular oxygen concentration in the solution must be low.
In this work, we used acetonitrile, which at a concentration of 0.1 M in aqueous solutions causes a full reaction to form mainly [Cu (CH3CN)2]+(aq) complexes [13].
In biological systems, the possibility of linking monovalent copper to amino groups (proteins, nucleic acids), organic sulfur (methionine, cysteine, cystine), and unsaturated or aromatic systems should not be overlooked [17-20].
ATP forms a complex with copper ions (Cu2+ and Cu+ ions; the latter is more stable) in which the metal ion is bound both to the nitrogen base and to the negatively charged oxygen in the phosphate groups, forming an unusually distorted complex (Figure 10) [16,21,22]. It appears that these complexes can inhibit enzymatic systems that are dependent on ATP.
The goal of this paper is to measure the effect of Cu+ on multiple enzymatic systems, and advance our understanding of the mechanism of its biocidal activity. The systems include polymerase (studied using the PCR system); the restriction enzyme EcoP15I, a type 3; a T4 ligase that requires ATP; EcoRI, a type 2; alpha and beta amylase, which don’t require ATP; and phosphates that operate on the phosphodiester bond, studied via standard lab procedure.
The synthesis of DNA molecules from deoxyribose nucleotides is catalyzed by DNA polymerase [22]. DNA polymerase enzymes are essential for DNA replication, during which the DNA polymerase copies the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the following reaction:
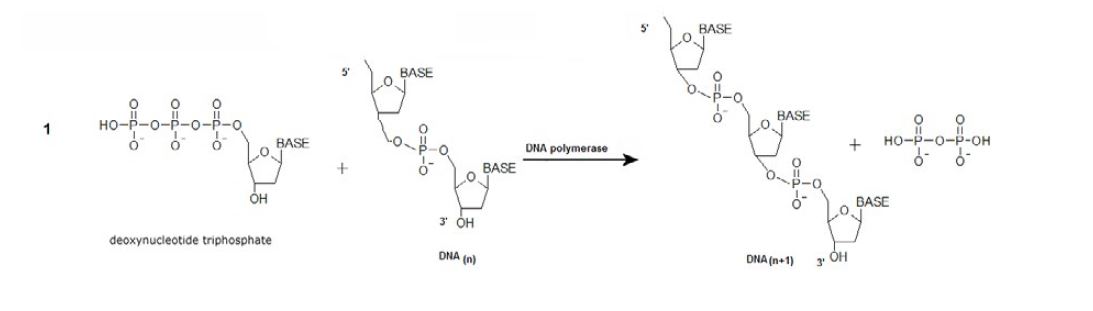
Equation 1
The polymerase chain reaction (PCR) is a commercial method, widely used nowadays in molecular and medical biology to make several (thousands to billions) copies of a specific DNA segment. The method uses a heatstable DNA polymerase, such as Taq polymerase, an enzyme originally isolated from thermophilic bacteria [23].
The polymerase chain reaction (PCR) is a commercial method, widely used nowadays in molecular and medical biology to make several (thousands to billions) copies of a specific DNA segment. The method uses a heatstable DNA polymerase, such as Taq polymerase, an enzyme originally isolated from thermophilic bacteria [23].
This study utilizes the commercial Polymerase Chain Reaction (PCR) system in anaerobic conditions.
The ligase T4 DNA enzyme facilitates the binding of DNA strands by catalyzing the formation of a phosphodiester bond (Equation 2).

Equation 2
The T4 ligase is one of the most commonly used enzymes in laboratory research. For our purposes, it should be noted that it requires ATP as a cofactor [24].
Restriction enzymes are enzymes that cleave DNA into fragments at or near specific recognition sites. There are three types of restriction enzymes: type 1 requires both ATP and S-adenosyl-L-methionine to function; type 2 does not require ATP; type 3 requires ATP, but does not hydrolyze it; and type 4 enzymes target modified DNA [25].
In this study, we used the restriction enzyme EcoP15I, a type 3 that requires ATP, and EcoRI, a type 2 that does not require ATP, operating on a pZIP plasmid.
Acid phosphatase (EC 3.1.3.2) is an enzyme used to free attached phosphoryl groups from other molecules. This enzyme occurs in many species of animals and plants and is essential for maintaining normal metabolic activity. This study used the para nitrophenyl phosphate as a substrate, obtaining para nitrophenol as a product [26] (Equation 3).
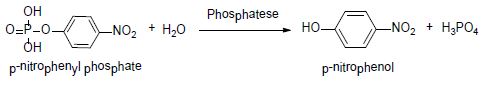
Equation 3
The system is easy to follow with UV-VIS spectroscopy via the 410nm absorption band of the product.
The enzyme α-amylase (EC 3.2.1.1) breaks down longchain saccharides (starch), yielding maltose, glucose, and “limit dextrin” from amylopectin (Equation 4) [27].
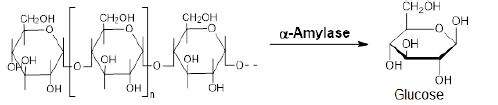
Equation 4
The enzyme β-amylase (EC 3.2.1.2 ) [21] (alternative names: 1,4-α-D-glucanmaltohydrolase; glycogenase; saccharogen amylase) is also synthesized by bacteria, fungi, and plants. Working from the non-reducing end, β-amylase catalyzes the hydrolysis of the second α-1,4glycosidic bond, cleaving off two glucose units (maltose) at a time (Equation 5).
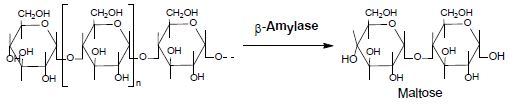
Equation 5
Enzyme reaction rates can be decreased by various types of enzyme inhibitors, namely competitive, non-competitive, uncompetitive, and irreversible. A kinetic study must be performed to determine the nature of the inhibition; such research has been performed on the phosphatase system since it is simpler for kinetic monitoring.
Materials and Methods
Anaerobic conditions (described in our previous studies [1,2]
Anaerobic medium: The anaerobic medium obtained by bubbling argon gas through the reaction medium. The reaction medium in most experiments kept in an airtight container, such as a bottle of penicillin or a syringe. In experiments where it was difficult to work in sealed containers (PCR, restriction enzymes EcoP15I and EcoRI, and T4 DNA Ligase enzyme), the reaction solution was kept under a stream of argon gas during the anaerobic stage.
Production of Cu+ ions in the experimental solution [15]: Prior to enzymatic experiments, Cu+ ions were produced from deaerated aqueous solutions containing a mixture of CuCl2 as the source for Cu2+ ions, metallic copper, and 0.1 M acetonitrile as a stabilizing ligand, according to equation 6:
Equation 6
Injection of copper ion solutions via syringes: Solutions containing Cu+ ions were gently injected through a three-way syringe valve into the anaerobic medium.
Gel electrophoresis: Gel electrophoresis was used to separate the DNA fragments based on their size and charge, in order to identify and quantify PCR, Ligase, and restriction enzymes’ activity under various experimental conditions. Quantification was performed by measuring the brightness level of the resulting stripe, and comparing it to the brightness level of a known fragment of a DNA marker (positive/negative control) in the same run. In order to obtain a normalized number indicating the activity of the enzyme, we used the “digital ImageJ” program (LOCI, The University of Wisconsin-Madison, research lab of Dr. Kevin Eliceiri, LABORATORY FOR OPTICAL AND COMPUTATIONAL INSTRUMENTATION). It should be noted that the quantification is for the activity of the enzyme, and not for the DNA fragments.
PCR (Polymerase chain reaction) [22]: The key ingredients of a PCR reaction are Taq polymerase, primers, template DNA, and nucleotides (DNA building blocks). The ingredients are assembled in a tube along with the cofactors required by the enzyme, and are subjected to repeated cycles of heating and cooling that allow the DNA to be synthesized.
In this study, all PCR experiments underwent the following stages: (1) 98˚C for 10 min; (2) 95˚C for 20 s; (3) 60˚C for 20 s; (4) 72˚C for 50 min; (5) Repetition of stages 2 to 4, for 35 times; (6) 72˚C for 5 min; (7) Cooling to 16˚C, until the samples were analyzed.
The PCR experiments used Normal Solution Composition (NSC), which contains the following: Taq polymerase, primers, PDNA, PCR water, and 0.2M dNTP (part of the ready mix solution). Most of the PCR experiments, in addition to the NSC solution, contained 0.3% CH3CN metal ions (as chloride salts), at final concentrations ranging from 80 to 0.08μM.
In the current study, we used as a template a 369bp DNA from an EGFP genome, produced from a pEPI-EGFP vector.
DNA durability to oxidation was measured on DNA segments (from plasmids cut by restriction enzymes) that were incubated with 8μM Cu2+, Ni2+, Zn2+, and Cu+ ions in an anaerobic atmosphere, at the PCR conditions described above. The segments were tested for changes using the Gel electrophoresis technique.
The T4 DNA Ligase enzyme (Sigma Aldrich Product No. D2886) was tested on three DNA segments produced from the plasmid pZIP-mCMV-RFP-Puro-(V033), isolated from E.coli bacteria, and cut by three restriction enzymes: EcoRI-HF, BamHI-HF, and Nhel-HF. The DNA segments were incubated with a ligase T4 enzyme for 24h at 16˚C with ATP and a buffer, Tris-HCl (procedure described in [27]) in the presence of Cu2+ and Cu+ ions (1% CH3CN), under anaerobic conditions. The resulting DNA, separated by electrophoresis, and compared to the reaction mixture without the enzyme.
Restriction enzymes EcoP15Iand EcoRI were tested on the plasmid pZIP-mCMV-RFP-Puro-(V033), isolated from E.coli using the recommended procedure, in the presence of Cu2+ and Cu+ ions (1% CH3CN), under anaerobic conditions. The resultant mixture was tested with the gel electrophoresis technique [28].
Acid Phosphatase (EC 3.1.3.2) was tested in the presence of Cu2+ and Cu+ ions (1% CH3CN), under anaerobic conditions on para nitrophenyl phosphate (PNPP) as a substrate, using a buffer (citric acid and ethylene di-amine) at pH 5.2, yielding para nitrophenol (PNP) as a product [25]. The product examined via UV-VIS spectroscopy at a 410nm absorption band at pH 9.5 against a calibration curve; a carbonate/bicarbonate buffer at pH 9.5 used to quench the enzymatic reaction.
The enzymes α-amylase (EC 3.2.1.1 ) (from Aspergillus oryzae) and β-amylase (EC 3.2.1.2 ) were tested on potato starch (sigma-aldrich), using a buffer of acetate, pH 4.8 [19]. The product (glucose in the case of α-amylase, and maltose in the case of β–amylase) quantities were determined with Nelson reagents, using CuSO4, which stoichiometrically oxidizes glucose and maltose at high pH values (carbonate/bicarbonate buffer pH 9.5) [29,30]. The reduced copper Cu2O with arsenomolybdate is determined via UV-VIS spectroscopy at a 650nm absorption band against a calibration curve. It is important to note that in an acidic environment, the divalent copper ion does not oxidize glucose or maltose, and the concentrations of copper ions in the enzymatic reaction are negligible relative to the concentrations used for the analytical determination of the sugar.
Processing Kinetic Results; from the dependence of the product on time, the maximum velocity of the reaction can be determined (Vmax), and from the dependence of the velocity of the reaction (V) on the substrate concentration ([S]), using Lineweaver-Burk equation [31] (Equation 7). It is possible to determine the Michaelis-Menten constant (Km), and whether the enzyme inhibition is competitive, noncompetitive, or uncompetitive.
Equation 7
Results
The effect of Cu+ ions on Taq polymerase (using the PCR system)
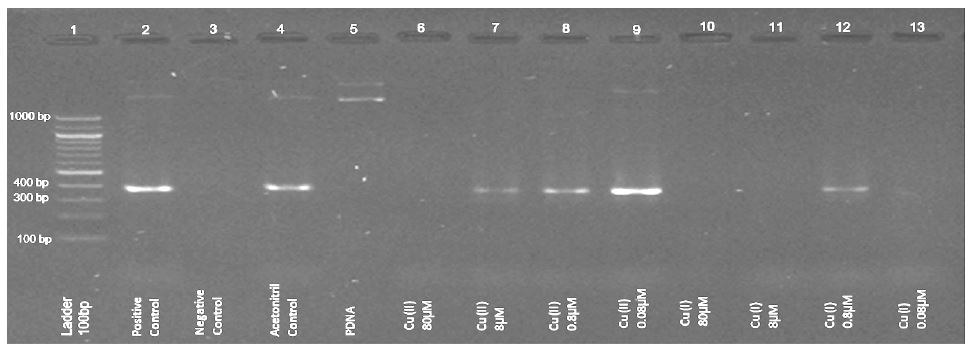
Comparison of the effect of copper ions’ concentrations on Taq polymerase under anaerobic conditions presented in figure 1 (electrophoresis results of PCR experiments).
The expected DNA product appears on 369bp (rows 2 and 4 in figure 1, positive and acetonitrile controls).
Cu2+ ions at a concentration of 1*10-4 M or higher inhibit DNA production. Cu+ ions, however, have a prominent effect: at concentrations of 1*10-7 M Cu+ ions (the lower limit of our capability), DNA production is inhibited (Figure 1, row 13).
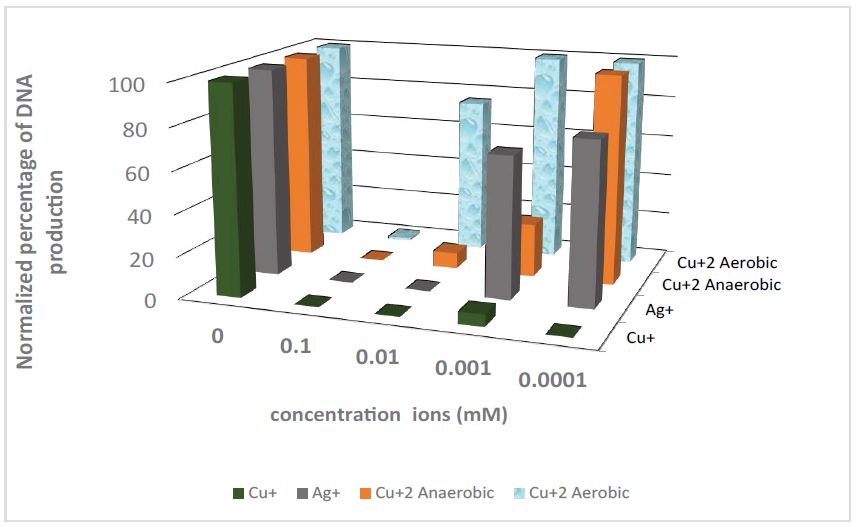
Figure 2 presents digital ImageJ processing normalized to the electrophoresis results of the PCR experiments under anaerobic conditions, for a range of Cu+, Ag+, Cu2+, and aerobic Cu2+ ions’ concentrations.
It is reasonable to assume that most of the Cu2+ ions’ effect is a consequence of partial reduction to Cu+ ions. In the aerobic environment, Cu+ ions oxidized very quickly to Cu2+ ions, reducing the Cu+ ions’ concentration; the results shown in figure 2 support this assumption. Moreover, the molecular oxygen reacting with Cu+ must produce reactive oxygen species (ROS), but it seems that the effect of the oxidative stress is negligible compared to the Cu+ effect.
Table 1 summarizes the relevant results of the PCR experiments using the Electrophoresis results analyzed with the ImageJ processing software. All experiments repeated at least twice.
Figure 2 and table 1 clearly indicate that Cu+ and Ag+ ions are different from Ni2+ and Zn2+: the latter has hardly any or no effect at all on Taq polymerase. It is shown that Cu+ ions block PCR in concentrations 100 times lower than Ag+.
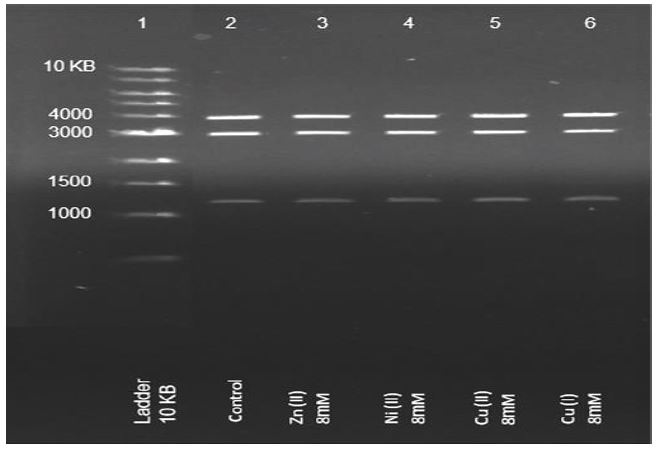
DNA durability to oxidation. To rule out DNA oxidation as the mechanism, DNA segments from plasmids cut by restriction enzymes (4,3 and 1.2KB) were incubated with 8μM Cu+2, Ni+2, Zn+2, and Cu+ ions under aerobic conditions. Figure 3 presents the electrophoresis results.
The results presented in figure 3 indicate that we can rule out DNA oxidation as the mechanism for the timescales and concentrations of the PCR experiments performed here.
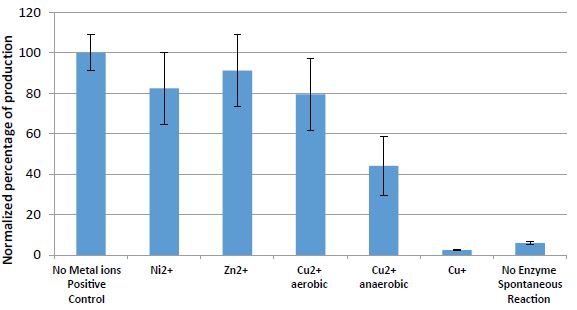
The effect of Ni2+, Zn2+, Cu2+, and Cu+ ions on phosphatase enzymes (from Aspergillus oryzae), operating on p-nitrophenyl phosphatase (PNPP) to produce p-nitrophenol (PNP) (Equation-3), is presented in figure 4.
Figure 1:Example of gel-electrophoresis results of PCR experiments under anaerobic conditions. Normal Solution Composition (NSC) with 0.3% CH3CN was used, with additions of Cu+2 and Cu+ ions at final concentrations between 100 and 0.1μM.
Figure 2: Gel-Electrophoresis results of PCR experiments (relative strength of the signal obtained by normalization with respect to the signal of the positive control). In anaerobic conditions, NSC with 0.3% CH3CN was used, with additions of Cu+, Ag+, Cu+2, and aerobic Cu+2 ions’ concentrations between 0.1 and 0.0001mM.
Figure 3: Gel-electrophoresis results of DNA segments (from plasmids cut by restriction enzymes) that were incubated with 8μM Cu+2, Ni+2, Zn+2, and Cu+ ions under aerobic conditions.
The results in figure 4 show the dramatic impact of Cu+ ions on the phosphatase enzyme.
It can be observed that the effect of bivalent copper, zinc, and nickel ions is negligible. However, under anaerobic conditions, bivalent copper ions have an effect that may contribute to the formation of monovalent copper ions.
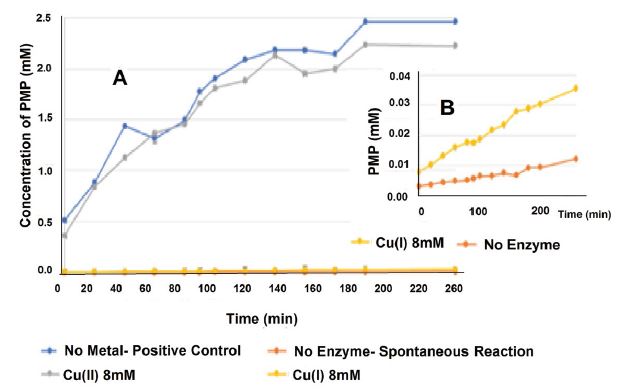
Figure 5 presents the kinetic results of the reaction in the presence of 8mM Cu2+ and Cu+ ions (1% CH3CN), under anaerobic conditions.
Table 1:Relevant results of PCR experiments.
| Experiment | Digital Normalized signal | Signal at 369bp | SD |
|---|---|---|---|
| Ladder | 10~ |  |
0 |
| NSC (Positive control) | 100 |  |
0 |
| NSC without Taq polymerase (Negative control) | 0 |  |
0 |
| NSC +0.3% CH3CN(Acetonitrile control) | 97 |  |
7 |
| NSC +0.3% CH3CN + 0.8μM Cu2+ | 26 |  |
14 |
| NSC +0.3% CH3CN + 0.08μM Cu2+ | 108 |  |
25 |
| NSC +0.3% CH3CN + 0.8μM Cu+ | 6 |  |
13 |
| NSC +0.3% CH3CN + 0.08μM Cu+ | 0 |  |
0 |
| NSC +1% CH3CN + 8μMAg+ | 0 |  |
0 |
| NSC +1% CH3CN + 0. 8μMAg+ | 68 |  |
6 |
| NSC +0.3% CH3CN +0. 8μM Ni+2 | 83 |  |
9 |
| NSC +0.3% CH3CN + 0. 8μMZn+2 | 100 |  |
1 |
| *NSC -Normal solution composition: Ready mix, Taq polymerase, primers, PDNA, PCR, and water. | |||
Table 2:Kinetic results of the effect of the phosphatase enzyme on the hydrolysis of the phosphate group from p-nitrophenyl phosphatase (PNPP) to p-nitrophenol (PNP).
| Km [mM] | ||
|---|---|---|
| 0.27 | 0.44 | No metal, Positive control |
| 0.21 | 0.31 | Cu+2 |
| 0.13 | 0.43 | Zn+2 |
| 0.23 | 0.39 | Ni+2 |
| 0.28 | 0.17 | Cu+ |
Figure 4: Results (normalized percentage of PNP) of the operation of the phosphatase enzyme on the hydrolysis of the phosphate group from p-nitrophenyl phosphatase (PNPP) to p-nitrophenol (PNP) in the presence of 10mM Cu2+ (aerobic and anaerobic), Ni2+, Zn2+, and Cu+ ions (1% CH3CN) under anaerobic conditions, pH 5, 37 oC, for 90min.
The results in figure 5 demonstrate the dramatic impact of Cu+ ions on the phosphatase enzyme.
To determine the nature of the inhibition, kinetic experiments were performed at several substrate concentrations (0.1-20mM) in the presence of Zn2+, Ni2+, Cu2+, and Cu+ ions (1% CH3CN), under anaerobic conditions
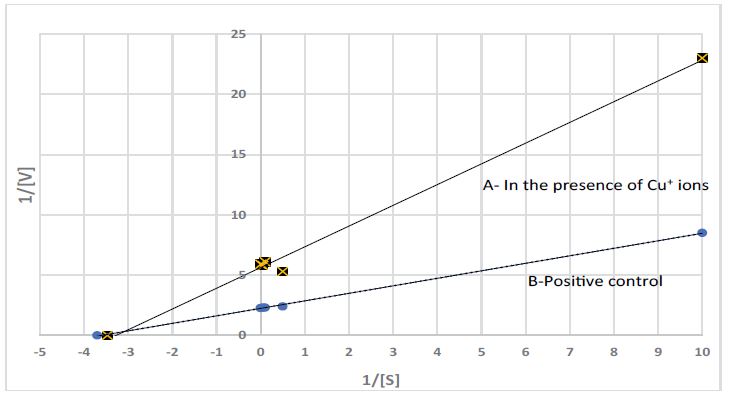
A Lineweaver-Burk plot was used for determination of Vmax and Km; the plot is shown in figure 6 and the values (Vmax and Km) are presented in table 2.
From the Lineweaver-Burk plot and from Vmax and Km values it is clear that Cu+ ions are non-competitive inhibitors to the phosphatase enzyme in the (PNPP) /(PNP) system.
Figure 5:Kinetic results of the operation of the phosphatase enzyme on the hydrolysis of the phosphate group from p-nitrophenyl phosphatase (PNPP) top-nitrophenol (PNP) in the presence of 8mM Cu2+ and Cu+ ions (1% CH3CN), under anaerobic conditions. A: An overview of the kinetics. B: A zoom-in on the kinetics of Cu+ ions, compared with the spontaneous reaction.
Figure 6:Lineweaver-Burk plot of the phosphatase enzyme operation on the hydrolysis of the phosphate group from p-nitrophenyl phosphatase (PNPP) top-nitrophenol (PNP) A- in the presence of Cu+ ions; B- Positive control, (1% CH3CN), under anaerobic conditions and substrate concentrations ranging from 0.1 to 20mM.
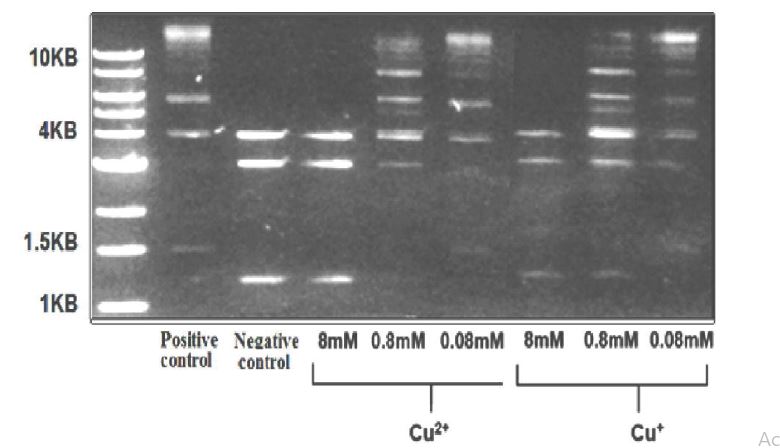
The effect of Cu+ ions on Ligase T4 DNA: Figure 7 presents electrophoresis results of the ligase T4 DNA enzyme operating on 3 DNA segments (4,3, and 1.2 KBnegative control) producing longer DNA segments (1.5,4.1,6, and larger than 10 KB- positive control). These experiments were performed under anaerobic conditions in the presence of Cu+2 and Cu+ ions.
The results in figure 7 show that while Cu+ and Cu2+ ions both inhibit the enzyme, the effect of Cu+ ions is more substantial (the signal of the 1.2KB DNA segment is still apparent with 0.8 mM Cu+ ions).
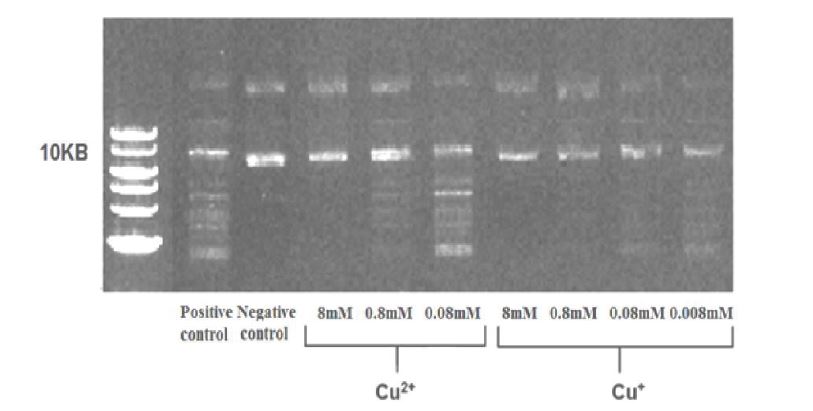
The effect of Cu+ ions on the restriction enzyme EcoP15I: Fig. 8 presents the electrophoresis results of EcoP15I enzymes operating on pZIP plasmids (negative control), producing DNA segments (positive control).
Figure 8 shows that Cu+ and Cu2+ ions both inhibit the restriction enzyme EcoP15I. The DNA segment remains at the same position as the negative control (10KB), and no other DNA segments appear as a result of the enzyme’s action, as shown by the positive control (>10KB). In addition, the effect of Cu+ is more substantial (inhibition above 0.1mM).
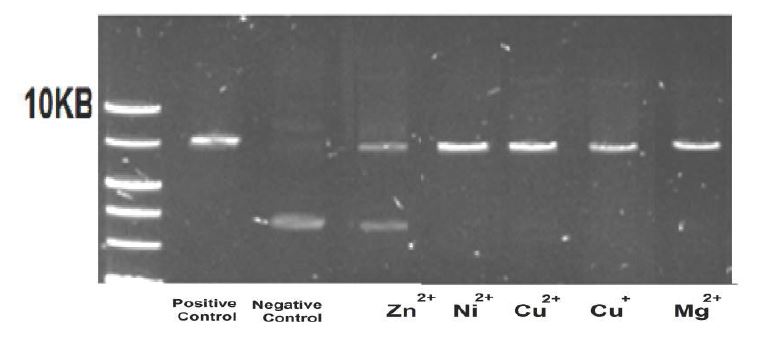
The effect of Cu+ ions on the restriction enzyme EcoRI: Figure 9 presents the electrophoresis results of the EcoRI enzyme operating on pZIP plasmids (negative control; the pZIP plasmids are still closed circles, a signal appears at ~5KB), producing DNA segments (positive control; the enzyme is cut in one place, opening the plasmid, and resulting in a signal at distances as low as 10KB). The experiments were performed under anaerobic conditions in the presence of 0.8mM Zn2+, Ni2+, Cu2+, Cu+, and Mg2+ ions (Mg2+ above the controls).
The results in Fig. 9 show no effect of Cu+ and Cu2+ ions on the EcoRI enzyme.
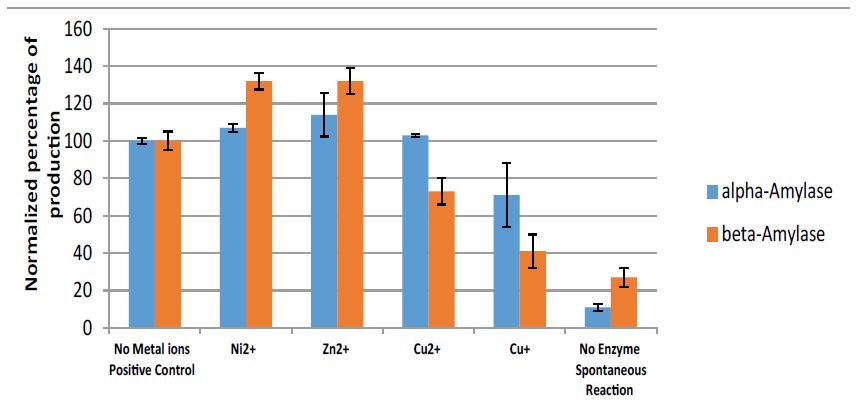
Effect of Cu(I) ions on α-Amylase and β-Amylase: The effect of 10mM Ni2+, Zn2+, Cu2+, and Cu+ ions on α amylase and β-amylase enzymes, operating on starch to produce glucose and maltose, respectively, is presented in figure 10.
Figure 7:Electrophoresis results of DNA segments (from plasmids cut by restriction enzymes) that were incubated with Ligase T4 enzymes for 24h at 16 oC,in the presence of Cu+2 and Cu+ ions (1% CH3CN), under semi-aerobic conditions.
Figure 8:Electrophoresis results of EcoP15I enzymes operating on pZIP plasmids in the presence of Cu2+ and Cu+ ions (1%CH3CN).
Figure 9:Electrophoresis results of the EcoRI enzyme operating on pZIP plasmids in the presence of 0.8mM Zn2+, Ni2+, Cu2+, Cu+, and Mg2+ ions (1% CH3CN), under anaerobic conditions.
Figure 10:alpha-amylase enzyme (blue): Results (normalized percentage of glucose) for 3% starch in the presence of 10mM Cu2+, Ni2+, Zn2+, and Cu+ ions (1%CH3CN) under anaerobic conditions, pH 5, 40oC, for 60min. beta-amylase enzyme (red): Results for 3% starch in the presence of 10mM Zn2+, Ni2+, Cu2+, and Cu+ ions (1% CH3CN) under anaerobic conditions, pH 4.8, 50oC, for 24h.
From the results presented in figure 10, we can conclude that there is an inhibition effect caused by Cu+ ions compared to zinc and nickel ions, and the effect is more pronounced in the case of beta amylase.
Discussion
In this study, we attempted to demonstrate the significant effect that monovalent copper ions have on easy-to-monitor enzymatic systems.
The first groups of enzymes are DNA Taq polymerase enzymes, DNA-restriction and ligation enzymes, all monitored by electrophoreses.
The Taq polymerase enzyme measured using a commercial PCR system at elevated temperatures, thus preventing atmospheric oxygen dissolution, and enabling the maintenance of a highly effective anaerobic solution. The results show dramatic inhibition, exerted by monovalent copper ions, where using less than 1*10-7 M of Cu+ ions the system is completely shut down, while Ag+ ions required 1*10-5 M, and 1*10-5 M of Cu2+ ions exhibited only a minor effect.
figure 2 clearly shows that maintaining anaerobic conditions allows Cu+ to inhibit the Taq polymerase in all ranges of concentration investigated here. Allowing molecular oxygen to interfere decreases the effect of copper ions. The effect of divalent copper can be partially attributed to the presence of monovalent copper, whose ratio increases under anaerobic conditions [32,33].
T4 DNA ligase enzyme and type 3 restriction enzyme EcoP15I are significantly inhibited by copper ions. While Cu+ and Cu2+ ions both inhibit the enzymes, the effect of Cu+ ions is more substantial.
Unlike the abovementioned enzymes, which require ATP as a cofactor, the type II restriction enzyme EcoRI, which does not require ATP as a cofactor was not affected at all by copper ions in general, nor by monovalent copper ions in particular.
Out of the first group of enzymes tested (enzymes of restriction, ligation, and polymerization of DNA), those that require ATP were significantly inhibited by monovalent and divalent copper ions, while the one enzyme that does not require ATP was not affected at all by the copper ions.
The second group of enzymes, which includes phosphatase and alpha and beta amylase, were examined by spectrophotometry methods. These enzymes do not require ATP to function, although the phosphatase enzyme does act on ATP, or derivatives similar to ATP.
The phosphatase enzyme was studied via hydrolysis of the phosphate group from p-nitro-phenyl- phosphatase (PNPP) to p-nitro-phenol (PNP), which is easy to monitor directly using 410 nm absorption band of the product. The system can be maintained at anaerobic conditions, which allow working with known concentrations of monovalent copper.
Phosphatase was found to be dramatically and noncompetitively inhibited by monovalent copper (Fig. 5). Its substrate’s aromatic ring partially resembles the adenosine in ATP. As with adenosine, monovalent copper interacts with the aromatic ring, and probably also with the negatively charged oxygen on the phosphate [15,34]. That interaction may be related to the inhibitory mechanism. Divalent copper does not interact with the aromatic ring, and does not inhibit the enzyme.
Monovalent copper ions were also found to inhibit both alpha and beta amylase enzymes. The effect is not as strong as on phosphatase, but is definitely significant.
The alpha and beta amylase enzymes do not need ATP to function. The fact that monovalent copper inhibits them clearly does not support the assumption that the complex of monovalent copper with ATP plays an important role in the enzyme-inhibiting mechanism. The effect of monovalent copper ions on these particular enzymes seems to result from some other mechanism; because amylase enzymes contain SS bonds, there is a basis for the assumption that monovalent copper break down the SS bond in proteins [35].
In this study, we showed that monovalent copper and monovalent silver ions (tested only in the PCR system) inhibit essential enzymes.
The mechanism is not one of oxidative stress, even though monovalent copper ions (much more than bivalent iron ions) react with molecular oxygen (that does not require hydrogen peroxide, as divalent copper and iron ions) to generate reactive oxygen species (ROS), so that a Fenton-like reaction can commence without hydrogen peroxide [4]. If the mechanism was one of oxidative stress, we would expect that an aerobic atmosphere would enhance the effect. However, the results show the opposite: an anaerobic atmosphere increases the effect. Copper ions, both monovalent and bivalent, do indeed generate reactive oxygen species (ROS), but the impact of the latter on the systems is negligible compared to the main effect. Monovalent silver ions do not generate reactive oxygen species (ROS), and yet Ag+ inhibits the polymerase enzyme (and has a bactericidal effect), apparently with a mechanism similar to that of the Cu+.
Previous studies that have examined the role of oxidative stress generated by copper ions found no damage to DNA in vivo [7,8,9]. At non-cytotoxic concentrations, copper ions inhibit the repair of oxidative DNA damage induced by visible light, the inhibition probably resulting from damage to enzyme function caused by Cu+ ions, rather than from oxidative stress [8]. Figure 6 shows that DNA plasmids maintain their weight, and are not harmed by monovalent copper ions and molecular oxygen.
In fact, Cu+ ions can serve as an antioxidant agent. Recent results show that a Cu+ ion complex with ATP reacts very rapidly with methyl radicals (CH3.) to produce Cu2+ ions and methane, terminating the radical chain (Equation 8) [16].
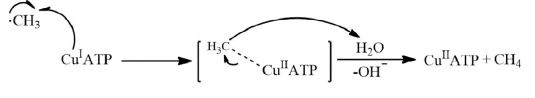
Equation 8
Indeed, a previous report showed that copper ions reduce the oxidative DNA damage caused by Fe2+ ions and H2O2 [8].
In our experiments, thanks to the acetonitrile in the medium, Cu+ ions do not undergo the disproportion reaction that is expected in aqueous solutions. Recent results show that ATP and adenosine form a strong complex with Cu+ ions, and shift the disproportion reaction to the left [16].
In vivo, it is reasonable to assume that ATP and other nucleic acids serve as stabilized ligands for Cu+ ions inside the cell.
Stronger affinity of Cu2+ ions to the nitrogen lone pair electrons of the adenine base interact with the negatively charged phosphate groups, resulting in a distortion of the ATP complex [21] (Figure 10).
Calculations suggest that the complex Cu+-ATP is similar to the Cu2+-ATP complex. The Cu+ ion interacts strongly with the base (adenine) and the to the phosphate groups causes the ATP to distort and fold, where the distorted complexes probably disrupt the function of ATP as a co-factor in the enzymatic systems [16].
Ions such Zn2+ and Ni2+ have a negligible effect on the system; Cu2+ has some effect (Figure 5). It is known that enzymatic systems require Mg2+ ions for proper operation of the enzyme, but it was not observed in the range of concentrations tested that the partial exchange of Mg2+ ions by Zn2+ and Ni2+ or Cu2+ had a significant effect. It is possible that most of the effect of Cu2+ is due to a reduction to Cu+ in a stabilized Cu+ environment. It was mentioned in the literature that Cu2+ ions are rapidly reduced to Cu+ ions by sulfhydryl ligands in solution, including glutathione and cysteine in the cytoplasm [33].
The assumption that the Cu+ ions’ anti-bacterial mechanism functions via enzymatic inhibition is also supported by recent results, showing that elevated temperatures boost the antibacterial action of Cu+ ions [2]. In less than 1 min at 40˚C, 0.4mM of Cu+ ions eliminated a 105 bacterial population; by comparison, an experiment at 20˚C left 102 bacteria surviving after 1 min, and Cu2+ ions at control conditions had no effect at all. Heat stimulates the metabolism, and permanent enzymatic inhibition is amplified by enzymatic activity. Furthermore, the result demonstrates that the antibacterial effect of Cu+ is higher when the E.coli utilizes glycerol as the sole carbon source, compared to lactose and glucose. Bacteria adapted to glycerol as the sole carbon source are more vulnerable to the impact of enzymatic inhibition than bacteria that grow with glucose. Apparently, the effect is not only via damage to cell division, since if that were the case, lengthening the generation time would minimize the effect. The results show the opposite.
Several previous studies have demonstrated the anticancer effects of copper oxide. In a recent paper, the Fedoped CuO NPs were tested in vivo, resulting in complete tumor remission in multiple syngeneic subcutaneous mouse models. In cancerous cells, the metabolic rate is much larger than in normal cells. It is therefore likely to be much more susceptible to the inhibitory effect of copper ions, or more precisely to Cu+ ions. It seems that the Cu2+ ions from the CuO NPs, reduced by glutathione or other reducing agents to Cu+ ions in the cancerous cells, are probably inhibiting the DNA polymerase.
We suggest that Cu+ and Ag+ ions act by interfering with enzymatic metabolisms, such as DNA polymerase, phosphatase, etc., via uncompetitive inhibition of the enzymes.
More relevant than ever, this finding may suggest a possible role of Cu+ ions producing systems as antiviral agents as well. However, that concept is yet to be proven.
Conclusion
Monovalent copper ions inhibit enzymes essential for cell function. This can explain the significant antibacterial effect of monovalent copper, an explanation that has been missing in the literature for understanding “Copper tolerance and virulence in bacteria” [36,37].
Possible explanations for the mechanisms of bacterial copper toxicity included an oxidation mechanism, although monovalent copper ions react rapidly with molecular oxygen to obtain oxidative products (ROS). The oxidation mechanism, according to the findings of this paper, is negligible.
Another mechanism mentioned in the literature includes the breakdown of the SS bond in proteins [35]. However, except in the case of α and β amylase, this explanation is not valid for the cases examined in this study, as they do not contain any SS bonds [38,39].
It is known that in a biological enzymatic system, the monovalent copper ion can form a complex with different functional groups (the monovalent copper ion must bind to a stabilizing ligand in an aqueous environment to prevent the disproportionation reaction), there is a basis for considering that ATP plays an important role in stabilizing monovalent copper ions, and that the Cu+-ATP and Cu2+-ATP complexes have a role in the inhibition mechanism for ATP dependent enzymes.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Saphier M, Silberstein E, Shotland Y, Popov S, Saphier O (2017) Prevalence of Monovalent Copper Over Divalent in Killing Escherichia coli and Staphylococcus aureus. Current Microbiology 75: 426-430. [ Ref ]
Popov S, Saphier O, Popov M, Shenker M, Entus S, et al. (2019) Factors Enhancing the Antibacterial Efect of Monovalent Copper Ions. Current Microbiology 77: 361-368. [ Ref ]
Maillard JY, Hartemann P (2013) Silver as an antimicrobial: facts and gaps in knowledge. Critical Reviews in Microbiology 39: 373-383. [ Ref ]
Masarwa Md, Cohen H, Meyerstein D, Hickman DL, Bakac A, et al. (1988) Reactions of low-valent transition-metal complexes with hydrogen peroxide J Am Chem Soc 110: 4293-4297. [ Ref ]
Cervantes-Cervantes MP, Calderón-Salinas JV, Albores A, Muñoz- Sánchez JL (2005) Copper increases the damage to DNA and proteins caused by reactive oxygenspecies. Biological Trace Element Research 103: 229-248. [ Ref ]
Molphy Z, Slator C, Chatgilialoglu C, Kellett A (2015) DNA oxidation profiles of copper phenanthrene chemical nucleases. Front Chem. [ Ref ]
Linder MC (2012) The relationship of copper to DNA damage and damage prevention in humans. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 733: 83-91. [ Ref ]
Schwerdtle T, Hamann I, Jahnke G, Walter I, Richter C, et al. (2007) Impact of copper on the induction and repair of oxidative DNA damage”,Molecular Nutrition and Food Research 51: 201-210. [ Ref ]
Macomber L, Rensing C, Imlay JA (2007) Intracellular Copper Does Not Catalyze the Formation of Oxidative DNA Damage in Escherichia coli. American Society for Microbiology Journals 189:1616-1626. [ Ref ]
Rensing C, Fan B, Sharma R, Mitra B, Rosen BP (2000) CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci USA 97: 652-656. [ Ref ]
Grass G, Rensing C (2001) CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem Biophys ResCommun 286: 902-908. [ Ref ]
Tarrant E, P Riboldi G, McIlvin MR, Stevenson J, Barwinska-Sendra A, (2018) Copper stress in Staphylococcus aureus leads to adaptive changes in central carbon metabolism. Metallomics The Royal Society of Chemistry 11:183-200. [ Ref ]
Parker AJ, Macleod ID, Singh P (1981) Electrochemistry of copper in aqueous acetonitrile. J Solut Chem 10: 757-774. [ Ref ]
Kamau P, Jordan RB (2001) Complex Formation Constants for the Aqueous Copper(I)−Acetonitrile System by a Simple General Method. Inorg Chem 40: 3879-3883. [ Ref ]
Saphier M, Burg A, Sheps S, Cohen H, Meyerstein D (1999) Complexes of copper (I) with aromatic compounds in aqueous solutions. J Chem Soc Dalton Ttans. [ Ref ]
Lerner A, Meyerstein D, Blahman A, Saphier M, Yardeni G (2022) On the reactions of Cu(II/I)ATP complexes with methyl radicals. Journal of Inorganic Biochemistry Volume 234: 111883. [ Ref ]
Rubino JT, Chenkin MP, Keller M, Riggs-Gelasco P, Franz KJ (2011) A comparison of methionine, histidine and cysteine in copper(I)-binding peptides reveals differences relevant to copper uptake by organisms in diverse environments. Metallomics 3: 61-73. [ Ref ]
Rubino JT, Franz KJ (2012) Coordination chemistry of copper proteins: how nature handles a toxic cargo for essential function. J Inorg Biochem. 107: 129-143. [ Ref ]
Stamp L, Dieck HT (1987) Copper(I) complexes with unsaturated nitrogen ligands. Part III. Copper(I) diazadiene complexes with carbon monoxide, olefins and acetylenes. Inorganica Chimica ActaVolume 129: 107-114. [ Ref ]
Dome´nech A (2000) Electrochemistry of copper complexes with polyaza[n] paracyclophanes. Influence of ATP as an exogen ligand on the relative stability of the Cu(II) and Cu(I) oxidation states. Inorganica Chimica Acta 299: 238-246. [ Ref ]
Onori G (1987) A spectrophotometric study of the binding of Cu(II) ions to ATP. Biophysical Chemistry 28: 183-190. [ Ref ]
Chien A, Edgar DB, Trela JM (1976) Deoxyribonucleic acid polymerase from the extreme thermophile Thermusaquaticus. Journal of Bacteriology 127: 1550-1557. [ Ref ]
Lorenz TC (2012) Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. JoVE (Journal of Visualized Experiments) 2012: e3998. [ Ref ]
Ligases. Enzyme Resources Guide. Promega Corporation. [ Ref ]
Wyszomirski KH, Curth U, Alves J, Mackeldanz P, Möncke-Buchner E (2012) Type III restriction endonuclease EcoP15I is a heterotrimeric complex containing one Res subunit with several DNA-binding regions and ATPase activity. Nucleic Acids Res 40: 3610-3622. [ Ref ]
Berg JM, Tymoczko JL, Gatto GL (2001) Biochemistry. New York. [ Ref ]
Kundu AK, Das S (1970) Production of amylase in liquid culture by a strain of Aspergillus oryzae. Applied microbiology 19: 598-603. [ Ref ]
Thermo-Fisher Scientific. User Guide: Control Reaction for T4 DNA Ligase Activity. [ Ref ]
Sistla S, Rao DN (2004) S-Adenosyl-L-methionine-dependent restriction enzymes. Critical Reviews in Biochemistry and Molecular Biology 39:1-19. [ Ref ]
Cochran B, Lunday D, Miskevich F (2008) Kinetic analysis of amylase using quantitative Benedict’s and iodine starch reagents. Journal of chemical education 85: 401. [ Ref ]
Fleischer H (2019) The Iodine Test for Reducing Sugars–A Safe, Quick and Easy Alternative to Copper (II) and Silver (I) Based Reagents. World Journal of Chemical Education 7: 45. [ Ref ]
Hans L, Dean B (1934) The Determination of Enzyme Dissociation Constants”. Journal of the American Chemical Society 56: 658-666. [ Ref ]
Gorren ACF, Schrammel A, Schmidt K, Mayr B (1996) Decomposition of S-nitrosoglutathione in the presence of copper ions and glutathione. Arch Biochim Biophys 330: 219-228. [ Ref ]
Saphier M, Levitsky I, Masarwa A, Saphier O (2018) Complexes of copper(I) with aromatic compounds facilitate selective electrophilic aromatic substitution. Journal of Coordination Chemistry 71: 1738-1748. [ Ref ]
Nomura K, Yoneda I, Nanmori T, Shinke R, Morita Y, et al (1995) The Role of SH and S-S Groups in Bacillus cereus β-Amylase. The Journal of Biochemistry 118: 1124-1130. [ Ref ]
Ladomersky E, Petris MJ (2015) Copper tolerance and virulence in bacteria. Metallomics 7: 957-964. [ Ref ]
Freinbichler W, Colivicchi MA, Stefanini C, Bianchi L, Ballini C, et al. (2011) Highly reactive oxygen species: detection, formation, and possible functions. Cell Mol Life Sci 68: 2067-2079. [ Ref ]
Yoshida Y, Furuta S, Niki E (1993) Effects of metal chelating agents on the oxidation of lipids induced by copper and iron. Biochim Biophys Acta 1210: 81-88. [ Ref ]
Hiniker A, Collet JF, Bardwell JC (2005) Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J Biol Chem 280: 33785-33791. [ Ref ]