Journal Name: Journal of Applied Microbiological Research
Article Type: Review
Received date: 28 April, 2023
Accepted date: 26 May, 2023
Published date: 02 June, 2023
Citation: Marechal W, Stewart W, Gakpetor G, Badisa VLD et al. (2023) Review on the Bacterial-Mediated Methylmercury Formation in the Environment and Remediation. J Appl Microb Res. Vol: 6 Issu: 1 (20-28).
Copyright: 2023 Marechal W et al. This is an openaccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Methylmercury (MeHg) is highly toxic form of mercury (Hg) and causes neurotoxicity in humans. Its production in the environment is enhanced due to human activities such as massive industrialization and warmer temperatures which facilitate the activities of microbial methylators. It bioaccumulates mainly in seafood items and threatens human health. So far, review papers were mainly focused on MeHg toxicity and controlling soil conditions or soil amendment compounds to reduce MeHg formation. However, bioremediation plays an important role in the remediation of the metals in the environmental samples. Less attention has been paid to MeHg degrading bacteria that can control MeHg pollution. Therefore, to highlight current research, this review paper mainly focused on MeHg formation, the environmental conditions to reduce its formation in the environment, natural MeHg remediation, and experimentally developed bacteria for MeHg remediation.
Keywords
Methylmercury, Anaerobic bacteria, Demethylation, Safer environment.
Abstract
Methylmercury (MeHg) is highly toxic form of mercury (Hg) and causes neurotoxicity in humans. Its production in the environment is enhanced due to human activities such as massive industrialization and warmer temperatures which facilitate the activities of microbial methylators. It bioaccumulates mainly in seafood items and threatens human health. So far, review papers were mainly focused on MeHg toxicity and controlling soil conditions or soil amendment compounds to reduce MeHg formation. However, bioremediation plays an important role in the remediation of the metals in the environmental samples. Less attention has been paid to MeHg degrading bacteria that can control MeHg pollution. Therefore, to highlight current research, this review paper mainly focused on MeHg formation, the environmental conditions to reduce its formation in the environment, natural MeHg remediation, and experimentally developed bacteria for MeHg remediation.
Keywords
Methylmercury, Anaerobic bacteria, Demethylation, Safer environment.
Introduction
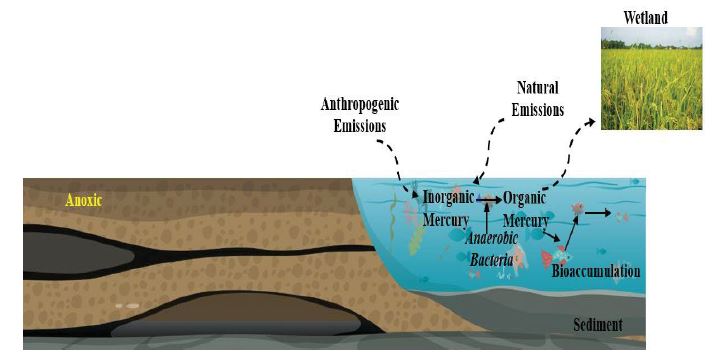
Mercury (Hg) is reported as a top three priority pollutant by the United States Environment Protection Agency (US EPA) and has been identified by the World Health Organization (WHO) as one of the ‘’ten leading chemicals of concern’’ [1-3]. The elemental (Hg0) or inorganic (Hg2+) form of Hg released into the environment from various natural or anthropogenic sources is less toxic to humans [4- 7]. However, these forms are converted to highly toxic compound methylmercury (MeHg) by anaerobic bacteria such as sulfate-reducing bacteria (SRB) and remain in the environment for several days. Consequently, it is accumulated and magnified in the food substances such as fish affecting humans and aquatic animals [8-11].
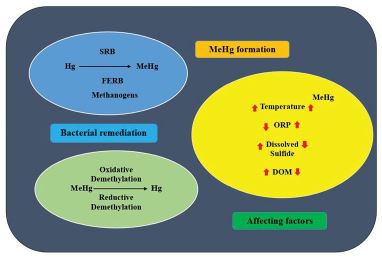
Figure 1:Graphical representation of abstract.
MeHg toxicity was first reported in Minamata City, Japan, affecting over 2500 people in the 1950s [12,13]. Fishermen and their families were the most affected people who ate fish daily [12]. The Minamata disease (MD) was first recognized as a mysterious neurological illness with severe uncontrollable tremors in Minamata in 1953 [14]. That disease was reported again between 1964 and 1965 in Niigata, near Tokyo [15,16]. The Japanese government authoritatively acknowledged that MeHg-containing seafood consumption was responsible for Minamata disease in 1968 [14]. Later, MeHg toxicity was also reported in other places like Ghana, Guatemala, Iraq, and Pakistan, due to flour consumption from wheat seeds treated with MeHg compounds [17].
It was reported that the MeHg accumulates in the fish or rice grains from the surrounding environment [18]. It was also reported that 75 to 90% of organic mercury exists as MeHg in those fish and shellfish [19,20]. It threatens the health of mainly seafood and rice lovers [21,22]. It was shown that people who eat fish regularly had increased total mercury levels in their hair than normal persons [23]. In the US, eating marine fish and shellfish is mainly responsible for MeHg intake in more than 90% of the population [24]. Hence, MeHg-contaminated fish is treated as the primary source of MeHg exposure to persons in the US. Americans take approximately 2.4 μg MeHg per week via fish, and a significant amount (2.3 μg) was absorbed into the body [25]. It was also reported that a significant US Gulf Coast population (30%) had higher MeHg concentrations in their blood because of eating MeHg-containing fish and developed neurodevelopmental problems in children [26]. Even in Florida Everglades for over three decades, Hg pollution had been a persistent concern due to elevated atmospheric Hg deposition, the system’s tendency for methylation, and rapid bioaccumulation. It was reported that a fetus, newborns, and children are at a higher health risk since they can have toxic effects even at low levels of MeHg exposure [27]. Based on a US birth cohort study. It was also reported that dental amalgams and seafood consumption d during pregnancy could cause respiratory infections in infants [28]. The maximum allowable daily Hg intake according to WHO and EPA was reported as 0.23 μg/ Kg/ day and 0.1 μg / Kg/day [29]. The half-life of MeHg in the human body was about 70 days, due to its slow removal and accumulation behavior in the body [30]. It was also reported that the inorganic mercury showed less toxicity in rats with a lethal dose (LD50) of 75 mg/kg, while MeHg showed higher toxicity in guinea pigs, mice, and rats with LD50 values of 21, 57.6, and 29.9 mg/ kg respectively [31-34]. Persons with 200–500 ng/mL Hg concentration in the blood or persons who ingest 3–7 μg Hg/ kg per day can show initial lethal effects of methylmercury [35]. Various health departments and Governments around the world have recognized the necessity for safeguard seafood to people; hence the highest safe ingestion limits for seafood were set as 0.46 ppm Hg and 1.6 μg MeHg/kg bodyweight as recommended weekly intake by the United States Food and Drug Agency (US FDA) [29].
It was reported that MeHg could bind to low molecular mass thiol proteins (LMM SH) like glutathione, high molecular mass proteins (HMM SH) such as albumin which contain sulfur or thiol-containing amino acids, and high molecular mass selenol (HMMSeH) proteins such as Glutathione peroxidase Px. [35,36]. It was also shown that it can also bind to nitrogen bases of DNA and RNA; however, the binding capacity is many times lesser than the thiol-containing proteins [37-44]. The exchange reactions between the MeHg-coupled LMM-SH and HMM-SeH proteins are responsible for the absorption, distribution, and excretion of MeHg in the human body [45-49]. The formation of MeHg coupled cysteine compound, Cys-S-HgMe, which can cross the cell membranes with the help of transporter L-type large neutral amino acid transporter (LAT1), change in antioxidant enzymes activity levels and reactive oxygen species production are mainly responsible for the MeHg toxicity in the humans [50-55]. It was shown that the nerve cells were more sensitive to MeHg than the glial cells since astrocytes contain less glutathione concentration than nerve cells [56]. It was reported that most of the MeHg (90-95%) from the ingested fish in humans was absorbed through the gastrointestinal tract and enters to the central nerves system [57]. It mainly affects central nervous system, and immune system of humans leading to visual impairment, tiredness, convulsions, paralysis of limbs, neurotoxicity, and can also cause death [58-66].
In the last two decades, much attention has been given to the bacterial bioremediation for cleaning polluted environment since it is easier, less time-consuming, and economically feasible than physical and chemical methods. The MeHg degrading bacteria were isolated from MeHgpolluted sites [67,68]. However, the degradation of MeHg has been much less studied so far [22]. Therefore, this review article mainly discusses MeHg formation, natural MeHg remediation, and experimentally developed bacterialmediated MeHg remediation.
Methylmercury Formation in the Environment
The inorganic Hg is converted into organic MeHg by various anaerobic bacteria through the methylation process in the environmental soil and water [69]. So far, 54 Hg methylating microorganisms were identified which comprises 37 sulfate reducing bacteria, 8 iron reducing bacteria, 8 methanogens, and 1 acetogenic microorganism, that contain the essential genes for methylation, hgcAB [10,69-72].
MeHg production in the environment depends on total Hg concentration, as well as several other environmental abiotic parameters like Hg speciation, pH, redox potential, temperature, microbial community, and inorganic as well as organic chelating agents [73]. Recently, a research study on the worldwide MeHg distribution and environmental factors of its production reported that MeHg concentration varied from 0.009 to 55.7 μg/kg at the different ecosystems, and the highest Hg methylation potential and MeHg concentration were found in paddy fields and marine environments, respectively (Table 1) [74].
In that study, the temperature (high temperature favors MeHg formation) and precipitation were recognized as important controllers of MeHg production [74]. It was also shown that oxidation-reduction potential (ORP) influences sulfur chemistry, thereby methylation of Hg. The Hg methylation is promoted by the microbial-mediated sulfur-reduction as a result of the decrease in ORP. The increased dissolved sulfide concentrations also decrease Hg methylation rates due to the removal of inorganic Hg as a sparingly soluble solid cinnabar or meta-cinnabar [69]. Hg can bind to the dissolved organic matter (DOM) and affect methylation by the methylating bacteria due to less availability of inorganic Hg for uptake since DOM molecules cannot cross the cell membrane of bacteria due to their large size [75]. The other abiotic factors, like humic and fulvic acids, were shown to play a role in Hg methylation [8,76]. Hg methylation particularly occurs in the floodplain soils rich in organic molecules due to their low oxygen conditions during flooding and organic substrates which serve as energy source for bacterial metabolism and sources for enhanced MeHg input to adjacent streams [77]. Recently, it was reported that up to 9% of Hg was converted to MeHg in the anaerobic setting in a study to know the input of Hg in urban runoff derived from historically contaminated soils and the subsequent production of MeHg in a stream–wetland complex (Durham, North Carolina) [78].
Table 1:Methylmercury levels at various environmental conditions.
| Environment | Sample description | MeHg level (ng/ L or μg/Kg)a | Location | Reference |
|---|---|---|---|---|
| Polar region | Snow | <0.02–0.03 | Antarctic | (Gionfriddo et al. 2016) |
| ≤0.015–0.118 | Canadian Arctic | (St. Louis et al. 2005, 2007) | ||
| Sea ice | <0.02–0.17 | Antarctic | (Gionfriddo et al. 2016) | |
| <0.02–0.57 | Arctic | (Beattie et al. 2014) | ||
| Sea water | N.A.b | Antarctic | (De Ferro et al. 2014) | |
| <0.02–0.15 | Antarctic | (Gionfriddo et al. 2016) | ||
| <0.01–0.18 | Southern Ocean | (Cossa et al. 2011) | ||
| 0.057–0.095 | Canadian Arctic | (St. Louis et al. 2007) | ||
| 0.015–178 | Canadian Arctic | (Kirk et al. 2008) | ||
| 0.021–0.126 | Arctic | (Wang et al. 2012) | ||
| Lake | Water | <0.085–0.257 | Antarctic | (Vandal et al. 1998) |
| 0.04–30 | Canadian Arctic | (Lehnherr et al. 2012a; Lehnherr et al. 2012b; St. Louis et al. 2005) | ||
| Sediment | 0.001–0.081 | Alaska, USA | (Poissant et al. 2008; Naidu et al. 2003) | |
| 0.26–3.4 | Alaska, USA | (Hammerschmidt et al. 2006) | ||
| 0.4–1.1 | Ny-Ålesund, Norway | (Jiang et al. 2011) | ||
| Soil | 0.01–<9.6 | Canadian Arctic | (Loseto et al. 2004; Oiffer and Siciliano 2009; St. Pierre 2015) | |
| Paddy fields | Non-contaminated soil | 0.02–1.76 | cMain rice planting areas, China | (Tang et al. 2019) |
| 0.84–4.5 | Chongqing, China | (Tang et al. 2018) | ||
| 0.17–1.0 | California, USA | (Tang et al. 2019) | ||
| 0.52–1.42 | California, USA | (Marvin-Dipasquale 2014) | ||
| 0.01–0.29 | Arkansas, USA | (Rothenberg et al. 2017) | ||
| Mining impacted area | Soil | 0.14–67 | Guizhou, China | (Rothenberg and Feng 2012; Li et al. 2019; Meng et al. 2010, 2014; Zhang et al. 2010a, 2010b) |
| 6.0–36.9 | Shaanxi, China | (Tang et al. 2018) | ||
| 2.8–10.9 | Hunan, China | (Meng et al. 2014) | ||
| 0.3–8.5d | Guangdong, China | (Meng et al. 2014) | ||
| aMeHg levels in snow, sea ice, and sea/lake water were represented in ng/L, while in sediment, wetland, and paddy fields soil was represented in μg/kg. bN.A. indicates data was not available. cMeHg level was measured in soil samples from 64 sites in 12 provinces in China, which accounts for 80% of the total rice planting area. dData from Pb/Zn mining impacted area. |
||||
Production of MeHg in the environment by microorganisms is shown in figure 1. Paddy fields, wetlands, lakes, and marine places which contain anaerobic conditions are most suitable for MeHg production [79]. The bacteria and extracellular polymeric substances (EPS) are mainly accountable for the production of MeHg and accumulate in those places, as shown in table 2. The transformed MeHg then accumulated into the food chain. Plants accumulate 104-105 times more MeHg than the surrounding waters [80]. It was shown that plants and animals contain MeHg approximately 1.0-6.5 μg/kg and 0.5-200 μg/kg [81,82]. It was also reported that the eatable clams, crabs, octopus, oysters, scallops, and squid in the US contain average THg concentrations ranging from 0.01 to 0.12 μg/kg wet weight (ww) [83]. The total Hg concentrations in terrapin scute and blood revealed that the organic form of Hg contributes to 90% of the total Hg [84]. In a recent study, it was showed that the altered total Hg and MeHg levels in rivers were important sources of MeHg to estuaries and coastal regions of the northern Gulf of Mexico (GOM) and were responsible for the increased levels of MeHg in GOM fish [85]. It was reported that in the Everglades, MeHg production mainly occurs at the periphyton region and varies with the season (3.5 g for the dry season and 37 g for the wet season) and contains six times more MeHg than the water (0.6 g for the dry season and 6.6 g for damp season) [17,86].
Table 2:Type of mercury methylators present in diverse environmental conditions.
| Environmental condition | Type of bacteria | Reference |
|---|---|---|
| Wetland sediments | SRB | (Bae et al. 2014) |
| FeRB | (Schaefer et al. 2014a, 2014b) | |
| Methanogens | (Bae et al. 2019) | |
| Syntrophs | (Christensen et al. 2019) | |
| Lake/river sediments | SRB | (Podar et al. 2015) |
| FeRB | (Bravo et al. 2018a) | |
| Methanogens | (Bravo et al. 2018b) | |
| Syntrophs | (Christensen et al. 2019) (Jones et al. 2019) (Yuan et al. 2019) | |
| Paddy soils | SRB | (Liu et al. 2014) |
| FeRB | (Liu et al. 2018) | |
| Methanogens | (Vishnivetskaya et al. 2018) | |
| Syntrophs | ||
| Forest soils | SRB | (Podar et al. 2015) |
| FeRB | (Xu et al. 2019) | |
| Methanogens | ||
| Syntrophs | ||
| Ocean | SRB | (Bouchet et al. 2018) |
| FeRB | ||
| Syntrophs | ||
| Marine conditions | SRB | (Podar et al. 2015) |
| FeRB | (Gionfriddo et al. 2016) | |
| Methanogens | ||
| Syntrophs | (Villar et al. 2020) | |
| Extreme environments | SRB | (Podar et al. 2015) |
| FeRB | (Christensen et al. 2019) | |
| Methanogens | ||
| Syntrophs | ||
| Bioreactor | SRB | (Podar et al. 2015) |
| FeRB | (Wang et al. 2019a, 2019b) | |
| Methanogens | ||
| Animal hindgut | Syntrophs | (Podar et al. 2015) |
| Abbreviations: SRB: sulfate-reducing bacteria; FeRB: iron-reducing bacteria | ||
Figure 2:Schematic representation of methylmercury formation in the natural environment.
Natural Methylmercury Remediation in the Environment
A potential strategy to decrease the MeHg levels in soil and water is determined by Hg methylation and MeHg degradation [87]. Recently, a study was conducted to determine whether specific carbon compounds affect the production potential of MeHg and methylating microbes’ distribution in those environmental samples [88]. As part of that study, sediment slurries were treated with alcohols, polysaccharides, or short-chain fatty acids. The results showed that lactate, ethanol, and methanol amendments had slightly increased MeHg, while cellobiose decreased MeHg production significantly (70%). Microbial communities were changed to non-hgcAB-containing Firmicutes (90%) in all the samples treated with cellobiose. These findings showed that simple methods could be used to decrease MeHg production in the environment [88]. In recently published reviews, there is a growing body of evidence that global and local perturbations influence Hg cycling and pollution management [22]. A wide range of soil composition factors determines the sorption, fate, and mobility of Hg in soils, including soil texture, organic matter content, hydroxides, and other organic and inorganic complexing agents determine how Hg is absorbed, the chemical form of Hg, pH, redox potential (EH), its fate as well as the specific stability of the bond between Hg and a ligand [77]. For the proper development of soil remediation techniques to effectively immobilize Hg by transforming it into stable and less toxic forms, knowledge of the above-mentioned factors is crucial [89]. Another method of reducing Hg mobility in soil is using soil amendments [90]. Organic amendments are particularly suitable since they show a high potential to immobilize Hg [90]. According to studies, Hg is usually bound to reduced sulfur functional groups (thiol, disulfide) of soil organic matter in an oxidized form such as Hg2+ [91]. It has already been demonstrated that Hg can be removed from solutions and combustion flue gases, reducing MeHg levels in rice grains, ad immobilizing MeHg in soil [90-93]. Researchers found that both biochar (BC) and Sugar beet factory lime (SBFL) treatment reduced the release of total Hg (Hgt) from the soil but not the methylation and ethylation of Hg [77]. There was also a report that Hgt, MeHg, and EtHg mobilization was generally higher at low redox potential and decreased as redox potential increased, regardless of soil treatment [77].
Various reports have shown that microorganisms adapt several metabolic pathways to survive in Hg/MeHg polluted environmental conditions [67,68]. The mer operon located on a plasmid or transposon or chromosome is responsible for the adaptation in Hg polluted environment [94]. The mer operon codes for MerR and MerD regulatory proteins, MerP, T, and E transport proteins, and MerA with reductase activity [76,95]. In response to Hg availability, the MerR or MerD regulatory protein binds at the promoter operator region and regulates the transcription of the MerA gene. During the bacterial Hg metabolic process, mercury ions are transported from the periplasm to the cytoplasm through transport proteins MerP/ MerD, and those ions are taken up by the mercuric reductase enzyme coded by MerA inside the cytoplasm. The enzyme reduces Hg2+ to mercury gas (Hg0) that diffuses passively out from the bacteria. Based on the mer determinants, Hg resistant bacteria are divided into broad and narrow ranges. The Hgresistant bacteria, which are limited range contain only the merA gene, while other bacteria, which are broad range, contain merB gene in addition to merA gene. The merB produces an organomercurial lyase enzyme and converts MeHg into inorganic mercury through the removal of methyl group [76].
Previous studies have shown that microbes degrade MeHg through oxidative demethylation or reductive demethylation process [76]. In anaerobic conditions, microbes degrade MeHg through the oxidative demethylation process; MeHg is converted to Hg2+ and carbon dioxide in that process. It was reported that the sulfur-reducing bacteria and methanogens are responsible for MeHg degradation in saturated soils through the oxidative demethylation process [76]. This oxidative demethylation process has also been observed in paddy fields with anaerobic conditions [96]. The research studies also revealed that the bacteria belonging to the Xanthomonadaceae family (Catenulisporaceae, Frankiaceae, Mycobacteriaceae, and Thermomonosporaceae) degrade MeHg in those paddy soils by demethylation pathway in the presence of Cu [97]. Furthermore, studies have also shown that Methylosinus trichosporium, an aerobic bacterium, degrades MeHg through an oxidative demethylation process, which is linked to Cu metabolic process [96]. In aerobic (oxic) environmental conditions such as watersaturated soils, the reductive demethylation process occurs, another MeHg degradation process. In that mechanism, the microorganisms contain mer operon coding for merB organomercurial lyase enzyme that degrades organic mercury to inorganic mercury, and merA reductase enzyme that reduce inorganic mercury to element Hg0 [98,99].
A recent report on world-wide photic and aphotic zones of oceans for MeHg degradation capacities through cultureindependent metagenomic and metatranscriptomic studies revealed that the capacity of biological MeHg degradation was extensively spread in the open ocean, and the highest capacity was observed in the mesopelagic zone [100]. It also revealed the presence of heterotrophic bacteria containing mere genes at different oceanographic regions and depths of open ocean, including polar regions. It was reported that Hg tolerance capacity depends on the bacterial strain, and a bacterium Alteromonas sp ISS312 unveiled a robust capacity of MeHg degradation that was isolated from South Atlantic Ocean bathypelagic water [100]. Recently, much focus was given to isolation of MeHg-degrading bacteria [101]. In that report, sixteen MeHg degrading bacteria were isolated from the contaminated wastewater sludge in Rio Grande do Sul, Brazil. It also showed that some isolates exhibited MeHg resistance to extreme concentrations of 8.7 μM. In that study, they also showed that the Pseudomonas putida V1 bacterium had only merA gene and converted the 90% of methylmercury in the medium to gaseous mercury. It was also reported that it has the ability to degrade MeHg under various pH (4-8) conditions, and temperatures (10–35°C), Pseudomonas putida V1 bacterium can grow even at the high concentration of 11.5 μM of MeHg [101]. Later, it was revealed that Pseudomonas putida V1 showed an alternative mechanism of MeHg degradation through the production of carbon dioxide during MeHg degradation, which did not involve merB product [101].
MeHg Bioremediation with Recombinant Technology using Bacteria
Hg and MeHg pollution can be controlled through bioremediation which is an easy, cost-effective and environmental-friendly approach than the physical or chemical methods. The usage of mer operon in Hg resistant bacteria is an attractive bioremediation approach for controlling Hg pollution. The mer operon occurs in different forms and locations in Hg-resistant bacteria. The MerB and MerA genes play an essential role in MeHg remediation efforts [102].
Recombinant plasmids were constructed with the cloning of some genes from the mer operon through Genetic engineering and introduced into the host bacteria, which were used to remove Hg from contaminated sites [103]. Other studies have focused on engineering bio-sorbent strains utilizing metal binding proteins or chelators such as metallothionein and polyphosphate kinase which play an essential role in binding the metals [104-107]. Biosorption is a passive process and hence microorganisms show limited metal binding capacity. In the Hg biosorption remediation process, specific methods are required to remove and recover Hg from the microorganisms. A recombinant E. coli strain containing merRTPAB genes was constructed for MeHg bioremediation and encapsulated in silica beads which act as a filtration material [107]. Following encapsulation, this strain also showed degradation of MeHg and exhibited the same degradation capacity as nonencapsulated cells [107]. Using recombinant microorganisms in the bioremediation process has certain limitations since runoff water from bioremediation can contain those unnatural bacteria, which can lead to a hazard [102]. In packed bed bioreactors, silica pumice granules are used to adsorb the natural mercontaining strains of Pseudomonas and it is the only method used till today to bioremediate and recover Hg at a technical scale [102]. Recently, MeHg-resistant Lactic acid bacteria (LAB) were isolated from feces (37) and breast milk (19) samples respectively from 19 volunteers in West Sekotong at, Indonesia which is an artisanal and small-scale gold mining site with high Hg levels. In the research studies, those bacteria showed different MeHg absorption abilities ranging from 17.375 to 51.597 mg/g of wet biomass after 24 h incubation. Out of those isolates, two bacteria isolated from the feces showed the highest Hg removal capacity and recognized as Enterococcus durans. The bacteria involved in MeHg remediation from all previous studies were summarized in table 3.
Table 3:Bacteria involved in MeHg remediation.
| Matrix | Type of bacteria | Removal efficiency | Reference |
|---|---|---|---|
| Soil | SRB and methanogens | - | (Barkay et al. 2003) |
| Soil | Methylosinus trichosporium | ~95% | (Lu et al. 2017) |
| Paddy soil | Catenulisporaceae, Frankiaceae, Mycobacteriaceae, and Thermomonosporaceae | > 75% | (Zhou et al. 2020) |
| South Atlantic Ocean | Alteromonas sp ISS312 | 98.2% | (Sanz-Sáez et al. 2022) |
| Sludge sewage from Rio Grande do Sul, Brazil | Pseudomonas putida V1 | 90% | (Cabral et al. 2016) |
| Waste site | Deinococcus radiourans | - | (Brim et al. 2000) |
| Wastewater | E.coli with mer-ppk fusion plasmid | >90% | (Kiyono et al. 2003) |
| Water | Enterococcus durans | > 70% | (Gasong et al. 2018) |
Perspectives and Recommendations
The recent global changes, such as increased anthropogenic activities with Hg and climate changes, can affect the microbial Hg methylation processes in Hg-contaminated ecosystems. Our knowledge of Hg methylators in a real environment is still limited and metagenomic analyses of Hg-contaminated ecosystems in the future can identify unknown species of Hg methylators that will enhance our knowledge of MeHg production in real environmental conditions [108]. In the future, metagenomic analysis of MeHg polluted environment should be carried out to identify better MeHg degrading bacteria that will help in the MeHg remediation process. Strict policies regulating Hg-related anthropogenic activities and adapting better remediation procedures can improve environmental and human health.
Conclusion
Environmental pollution due to natural and anthropogenic Hg emissions leading to the conversion of MeHg became a main risk to ecosystems and human health. The inorganic Hg is converted into organic MeHg by various anaerobic bacteria through the methylation process in the environmental soil and water. MeHg production in the environment depends on total Hg concentration and several other environmental abiotic parameters like Hg speciation, pH, redox potential, temperature, microbial community, and inorganic and organic chelating agents. A potential strategy to decrease the MeHg levels in the environment is determined by Hg methylation and MeHg degradation. MeHg degrading microorganisms contain MerB gene coding for organomercurial lyase enzyme that degrade MeHg to inorganic mercury and MerA gene coding for reductase, which converts to mercury gas Hg0. This review highlights that MeHg pollution can be controlled with bacterial bioremediation, which is an easy, cost-effective and environment-friendly approach.
Funding
No funding was involved for this paper since it is a review article that involved collecting research information.
Acknowledgments
The authors would like to acknowledge Ms. Rabi Elabor (School of the Environment, Florida A&M University) for the illustrated figure and Dr. Grisel Romero (School of the Environment, Florida A&M University) for her critical reading of the manuscript to enhance the quality of the manuscript.
Author Contributions
WM, WS, GG, VLDB, BM, and VI have perceived and planned the review. WM, WS, GG, and VLDB have contributed review material for original draft preparation. WM illustrated the figure. VLDB executed the editing. All authors approved the final version of this review paper.
Conflicts of Interest
The authors have declared that they do not have any conflict of interest.
Ethical Approval
This manuscript requires no ethical approval since it is a review paper.
Environmental Protection Agency (EPA) (2006) Mercury Compounds, Technology Transfer Network Air Toxics, Environmental Protection Agency (EPA). Washington, DC, USA. [ Ref ]
Spiegel SJ (2017) New mercury pollution threats: a global health caution. Lancet 390: 226-227. [ Ref ]
World Health Organization, (WHO) (2020) 10 chemicals of public health concern. [ Ref ]
Galbreath KC, Zygarlicke CJ (1996) Mercury speciation in coal combustion and gasification flue gases. Environ Sci technol 30: 2421- 2426. [ Ref ]
Tang T, Xu J, Lu R, Wo J, Xu X (2010) Enhanced Hg2+ removal and Hg0 re-emission control from wet fuel gas desulfurization liquors with additives. Fuel 89: 3613-3617. [ Ref ]
Sun M, Cheng G, Lu R, Tang T, Baig SA, et al. (2014) Characterization of Hg0 re-emission and Hg2+ leaching potential from flue gas desulfurization (FGD) gypsum. Fuel Process Technol 118: 28-33. [ Ref ]
Krzyżyńska R, Szeliga Z, Pilar L, Borovec K, Regucki P (2020) High mercury emission (both forms: Hg0 and Hg2+) from the wet scrubber in a full-scale lignite-fired power plant. Fuel 270: 117491. [ Ref ]
Weber JH (1993) Review of possible paths for abiotic methylation of mercury (II) in the aquatic environment. Chemosphere 26: 2063-2077. [ Ref ]
Kerin EJ, Gilmour CC, Roden E, Suzuki MT, Coates JD, et al. (2006) Mercury methylation by dissimilatory iron-reducing bacteria. Appl Environ Microbiol 72: 7919-7921. [ Ref ]
Parks JM, Johs A, Podar M, Bridou R, Hurt Jr RA, et al. (2013) The genetic basis for bacterial mercury methylation. Sci 339: 1332-1335. [ Ref ]
Villar E, Cabrol L, Heimbürger‐Boavida LE (2020) Widespread microbial mercury methylation genes in the global ocean. Environ Microbiol Rep 12: 277-287. [ Ref ]
Takeuchi T, Frank MD, Fischer PV, Annett CS, Okabe M (1977) The outbreak of Minamata disease (methyl mercury poisoning) in cats on Northwestern Ontario reserves. Environ Res 13: 215-228. [ Ref ]
Mergler D, Anderson HA, Chan LH, Mahaffey KR, Murray M, et al. (2007) Methylmercury exposure and health effects in humans: a worldwide concern. Ambio 36: 3-11. [ Ref ]
Eto K, Marumoto M, Takeya M (2010) The pathology of methylmercury poisoning (Minamata disease): The 50th Anniversary of Japanese Society of Neuropathology. Neuropathology 30: 471-479. [ Ref ]
Tsubaki T, Irukayama K (1977) Minamata disease. Methylmercury poisoning in Minamata and Niigata, Japan. The Quarterly Review of Biology 53. [ Ref ]
Eto K, Takahashi H, Kakita A, Tokunaga H, Yasutake A, et al. (2007) Pathological and biochemical studies of 30 Niigata autopsy cases related to Minamata disease. Nihon Eiseigaku zasshi 62: 70-88. [ Ref ]
Bae HS, Dierberg FE, Ogram A (2019) Periphyton and flocculent materials are important ecological compartments supporting abundant and diverse mercury methylator assemblages in the Florida Everglades. Appl Environ Microbiol 85: 1-17. [ Ref ]
Harada M (1995) Minamata disease-methylmercury poisoning in Japan caused by environmental-pollution. Crit Rev Toxicol 25: 1-24. [ Ref ]
Nam DH, Basu N (2011) Rapid methods to detect organic mercury and total selenium in biological samples. Chem Cent J 5: 3. [ Ref ]
Hong YS, Kim YM, Lee KE (2012) Methylmercury exposure and health effects. J Prev Med Public Health 45: 353-363. [ Ref ]
Paranjape AR, Hall BD (2017) Recent advances in the study of mercury methylation in aquatic systems. FACETS 2: 85-119. [ Ref ]
Hsu-Kim H, Eckley CS, Achá D, Feng X, Gilmour CC, et al. (2018) Challenges and opportunities for managing aquatic mercury pollution in altered landscapes. Ambio 47: 141-169. [ Ref ]
Mezghani-Chaari S, Hamza A, Hamza-Chaffai A (2011) Mercury contamination in human hair and some marine species from Sfax coasts of Tunisia: levels and risk assessment. Environ Monit Assess 180: 477- 487. [ Ref ]
Sunderland E, Mason R (2007) Human impact on ocean mercury concentration. Global Biogeochem Cycles 21: B4022. [ Ref ]
Mutter J, Curth A, Naumann J, Deth R, Walach H (2010) Does inorganic mercury play a role in Alzheimer’s disease? A systematic review and an integrated molecular mechanism. J Alzheimer’s Dis 22: 357-374. [ Ref ]
Evans DW, Cohen MD, Hammerschmidt C, Landing W, Rumbold D, et al. (2015) White Paper on Gulf of Mexico Mercury Fate and Transport: Applying Scientific Research to Reduce the Risk from Mercury in Gulf of Mexico Seafood. NOAA. Technical Memorandum NOS NCCOS. [ Ref ]
Feng L, Zhang C, Liu H, Li P, Hu X, et al. (2020) Impact of low-level mercury exposure on intelligence quotient in children via rice consumption. Ecotoxicol Environ S 202: 110870. [ Ref ]
Emeny RT, Korrick SA, Li Z, Nadeau K, Madan J, et al. (2019) Prenatal exposure to mercury in relation to infant infections and respiratory symptoms in the New Hampshire Birth Cohort Study. Environ Res 171: 523-529. [ Ref ]
World Health Organization, (WHO) (2008) Guidance for Identifying Populations at Risk from Mercury Exposure. WHO, Geneva, Switzerland. [ Ref ]
Miettinen JK (1973) Absorption and elimination of dietary mercury (2+) ion and methylmercury in man. In: Mercury, Mercurials, and Mercaptans. Proceedings 4th International Conference on Environmental Toxicology (Rochester), Plenum Press, New York. [ Ref ]
Young RAR (1992) Oak Ridge Reservation Environmental Restoration Program. [ Ref ]
Oriquat GA, Saleem TH, Naik RR, Moussa SZ, Al-gindy RM (2012) A Sub chronic Toxicity Study of Mercuric Chloride in the Rat. Jordan J Biol Sci 5. [ Ref ]
Jha A, Saidullah B, Bubber PA (2019) A study on prooxidative and neurotoxic effects of mercury chloride in rats. EC Pharmacol Toxicol 7: 175-187. [ Ref ]
Rai RK, Pati RS, Islam A, Roy G (2022) Detoxification of organomercurials by thiones and selones: A short review. Inorganica Chim Acta 538: 120980. [ Ref ]
Bjørklund G, Dadar M, Mutter J, Aaseth J (2017) The toxicology of mercury: Current research and emerging trends. Environ Res 159: 545- 554. [ Ref ]
Li Y, He B, Hu L, Huang X, Yun Z, et al. (2018) Characterization of mercury-binding proteins in human neuroblastoma SK-N-SH cells with immobilized metal affinity chromatography. Talanta 178: 811-817. [ Ref ]
Simpson RB (1964) Association constants of methylmercuric and mercuric ions with nucleosides. J Am Chem Soc 86: 2059-2065. [ Ref ]
Mansy S, Tobias RS (1974) Heavy metal-nucleotide interactions. II. Binding of methylmercury (II) to purine nucleosides and nucleotides studied by Raman difference spectroscopy. J Am Chem Soc 96: 6874- 6885. [ Ref ]
Mansy S, Tobias RS (1975) Heavy metal-nucleoside interactions. Binding of methylmercury (II) to inosine and catalysis of the isotopic exchange of the C-8 hydrogen studied by proton nuclear magnetic resonance and Raman difference spectrophotometry. Biochem 14: 2952-2961. [ Ref ]
Mansy S, Chu GY, Duncan RE, Tobias RS (1978) Heavy metal nucleotide interactions. 12. Competitive reactions in systems of four nucleotides with cis-or trans-diammineplatinum (II). Raman difference spectrophotometry of the relative nucleophilicity of guanosine, cytidine, adenosine, and uridine monophosphates and analogous DNA bases. J Am Chem Soc 100: 607-616. [ Ref ]
Prizant L, Olivier MJ, Rivest R, Beauchamp AL (1979) Metal binding to four different sites in adenine ligands. Crystal structures of 2:1 methylmercury complexes with adenine and 9-methyladenine. J Am Cheml Soc 101: 2765-2767. [ Ref ]
Hubert J, Beauchamp AL (1980) Metal binding to N (9), N (7), and N (3) of adenine. Crystal structure of μ-(adeninato-N 3, N 7, N 9) tris (methylmercury (II)) perchlorate. Can J Chem 58: 1439-1443. [ Ref ]
Charland JP, Beauchamp AL (1986) Preparation and structural studies of methylmercury compounds containing amino-deprotonated adenine. Inorg Chem 25: 4870-4876. [ Ref ]
Onyido I, Norris AR, Buncel E (2004) Biomolecule-mercury interactions: Modalities of DNA base− mercury binding mechanisms. remediation strategies. Chem Rev 104: 5911-5930. [ Ref ]
Fuhr BJ, Rabenstein DL (1973) Nuclear magnetic resonance studies of the solution chemistry of metal complexes. IX. Binding of cadmium, zinc, lead, and mercury by glutathione. J Am Chem Soc 95: 6944-6950. [ Ref ]
Rabenstein DL (1973) Nuclear magnetic resonance studies of the acidbase chemistry of amino acids and peptides. I. Microscopic ionization constants of glutathione and methylmercury-complexed glutathione. J Am Chem Soc 95: 2797-2803. [ Ref ]
Rabenstein DL, Evans CA (1978) The mobility of methylmercury in biological systems. Bioinorg Che 8: 107-114. [ Ref ]
Rabenstein DL, Reid RS (1984) Nuclear magnetic resonance studies of the solution chemistry of metal complexes. 20. Ligand-exchange kinetics of methylmercury (II)-thiol complexes. Inorgan Chem 23: 1246-1250. [ Ref ]
Arnold AP, Tan KS, Rabenstein DL (1986) Nuclear magnetic resonance studies of the solution chemistry of metal complexes. 23. Complexation of methylmercury by selenohydryl-containing amino acids and related molecules. Inorg Chem 25: 2433-2437. [ Ref ]
Aschner M, Aschner JL (1990) Mercury neurotoxicity: mechanisms of blood-brain barrier transport. Neurosci Biobehav Rev 14: 169-176. [ Ref ]
Kerper LE, Ballatori N, Clarkson TW (1992) Methylmercury transport across the blood-brain barrier. Am J Physiol Regul Integr Comp Physiol 262: 761-765. [ Ref ]
Yin Z, Jiang H, Syversen T, Rocha JB, Farina M, et al. (2008) The methylmercury-l-cysteine conjugate is a substrate for the L-type large neutral amino acid transporter. J Neurochem 107: 1083-1090. [ Ref ]
Farina M, Rocha JB, Aschner M (2011) Mechanisms of methylmercuryinduced neurotoxicity: evidence from experimental studies. Life Sci 89: 555-563. [ Ref ]
Lohren H, Bornhorst J, Galla HJ, Schwerdtle T (2015) The blood– cerebrospinal fluid barrier–first evidence for an active transport of organic mercury compounds out of the brain. Metallomics 7: 1420-1430. [ Ref ]
Branco V, Carvalho C (2019) The thioredoxin system as a target for mercury compounds. Biochim Biophys Acta (BBA)-Gen Subj 1863: 129255. [ Ref ]
Sanfeliu C, Sebastià J, Kim SU (2001) Methylmercury neurotoxicity in cultures of human neurons, astrocytes, neuroblastoma cells. Neurotoxicol 22: 317-327. [ Ref ]
Jinadasa BKKK, Fowler SW (2019) Critical review of mercury contamination in Sri Lankan fish and aquatic products. Mar Pollut Bull 149: 110526. [ Ref ]
Wakabayashi K, Kakita A, Sakamoto M, Su M, Iwanaga K, et al. (1995) Variability of brain lesions in rats administered methylmercury at various postnatal development phases. Brain Res 705: 267-272. [ Ref ]
Watanabe C, Satoh H (1996) Evolution of our understanding of methylmercury as a health threat. Environ Health Perspect 104(suppl 2): 367-379. [ Ref ]
Trasande L, Landrigan PJ, Schechter C (2005) Public health and economic consequences of methyl mercury toxicity to the developing brain. Environ Health Perspect 113: 590-596. [ Ref ]
Mutter J, Naumann J, Guethlin C (2007) Comments on the article “the toxicology of mercury and its chemical compounds” by Clarkson and Magos (2006). Crit Rev Toxicol 37: 537-549. [ Ref ]
Trasande L, Schechter CB, Haynes KA, Landrigan PJ (2006) Mental retardation and prenatal methylmercury toxicity. Am J Ind Med 49: 153- 158. [ Ref ]
da Silva DAF, Teixeira CT, Scarano WR, Favareto APA, Fernandez CD, et al. (2011) Effects of methylmercury on male reproductive functions in Wistar rats. Reprod Toxicol 31: 431-439. [ Ref ]
Basu N (2012) Piscivorous mammalian wildlife as sentinels of methylmercury exposure and neurotoxicity in humans. In Methylmercury and neurotoxicity. Springer, Boston 357-370. [ Ref ]
He X, Imanishi S, Sone H, Nagano R, Qin XY, et al. (2012) Effects of methylmercury exposure on neuronal differentiation of mouse and human embryonic stem cells. Toxicol Lett 212: 1-10. [ Ref ]
Nogara PA, Farina M, Aschner M, Rocha JB (2019) Mercury in our food. Chem Res Toxicol 32: 1459-1461. [ Ref ]
Mathema VB, Thakuri BC, Sillanpää, M (2011) Bacterial mer operonmediated detoxification of mercurial compounds: a short review. Arch Microbiol 193: 837-844. [ Ref ]
Agarwal M, Rathore RS, Jagoe C, Chauhan A (2019) Multiple Lines of Evidences Reveal Mechanisms Underpinning Mercury Resistance and Volatilization by Stenotrophomonas sp. MA5 Isolated from the Savannah River Site (SRS), USA. Cells 8: 309. [ Ref ]
Gilmour CC, Riedel GS, Riedel G, Kwon S, Landis R, et al. (2013) Activated carbon mitigates mercury and methylmercury bioavailability in contaminated sediments. Environ Sci Tech 47: 13001-13010. [ Ref ]
Gilmour CC, Bullock AL, Mcburney A, Podar M, Elias DA (2018) Robust mercury methylation across diverse methanogenic archaea. Mbio 9: e02403-e02417. [ Ref ]
Podar M, Gilmour CC, Brandt CC (2015) Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci Adv 1: e1500675. [ Ref ]
Yu R, Reinfelder JR, Hines ME, Barkay T (2013) Mercury methylation by the methanogen methanospirillum hungatei. Appl Environ Microbiol 79: 6325-6330. [ Ref ]
Ullrich SM, Tanton TW, Abdrashitova S (2001) Mercury in the aquatic environment: a review of factors affecting methylation. Crit Rev Environ Sci Technol 31: 241-293. [ Ref ]
Dai SS, Yang Z, Tong Y, Chen L, Liu SY, et al. (2021) Global distribution and environmental drivers of methylmercury production in sediments. J Hazard Mater 407: 124700. [ Ref ]
Mazrui NM, Jonsson S, Thota S, Zhao J, Mason RP (2016) Enhanced availability of mercury bound to dissolved organic matter for methylation in marine sediments. Geochimica et cosmochimica acta. 194: 153-162. [ Ref ]
Barkay T, Miller SM, Summers AO (2003) Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev 27: 355-384. [ Ref ]
Beckers F, Rinklebe J (2017) Cycling of Mercury in the Environment: Sources, Fate, and Human Health Implications - A Review. Crit Rev Environ Sci Tech 47: 693-794. [ Ref ]
Deonarine A, Hsu-Kim H, Zhang T, Cai Y, Richardson CJ (2015) Legacy source of mercury in an urban stream-wetland ecosystem in central North Carolina, USA. Chemosphere 138: 960-965. [ Ref ]
Tang W, Hintelmann H, Gu B, Feng X, Liu Y, et al. (2019) Increased methylmercury accumulation in rice after straw amendment. Environ Sci Technol 53: 6144-6153. [ Ref ]
Pickhardt PC, Fisher NS (2007) Accumulation of inorganic and methylmercury by freshwater phytoplankton in two contrasting water bodies. Environ Sci Technol 41: 125-131. [ Ref ]
Luengen AC, Flegal A (2009) Role of phytoplankton in mercury cycling in the San Francisco Bay Estuary. Limnol Oceanography 54: 23-40. [ Ref ]
Hammerschmidt CR, Fitzgerald WF, Lamborg CH, Balcom PH, Tseng CM (2006) Biogeochemical cycling of methylmercury in lakes and tundra watersheds of Arctic Alaska. Environ Sci Technol 40: 1204-1211. [ Ref ]
Yang L, Zhang Y, Wang F, Luo Z, Guo S, et al. (2020) Toxicity of mercury: Molecular evidence. Chemosphere 245: 125586. [ Ref ]
Blanvillain G, Schwenter JA, Day RD, Point D, Christopher SJ, et al. (2007) Diamondback terrapins, Malaclemys terrapin, as a sentinel species for monitoring mercury pollution of estuarine systems in South Carolina and Georgia, USA. Environ Toxicol Chem 26: 1441-1450. [ Ref ]
Buck CS, Hammerschmidt CR, Bowman KL, Gill GA, Landing WM (2015) Flux of Total Mercury and Methylmercury to the Northern Gulf of Mexico from U.S. Estuaries. Environ Sci Technol 49: 13992-13999. [ Ref ]
Li Y, Mao Y, Liu G, Tachiev G, Roelant D, et al. (2010) Degradation of methylmercury and its effects on mercury distribution and cycling in the Florida Everglades. Environ Sci Technol 44: 6661-6666. [ Ref ]
Kronberg RM, Jiskra M, Wiederhold JG, Björn E, Skyllberg U (2018) Corrections to Methyl Mercury Formation in Hillslope Soils of Boreal Forests: The Role of Forest Harvest and Anaerobic Microbes. Environ Sci Technol 52: 367. [ Ref ]
Christensen GA, Gionfriddo CM, King AJ, Moberly JG, Miller CL, et al. (2019) Determining the reliability of measuring mercury cycling gene abundance with correlations with mercury and methylmercury concentrations. Environ Sci Technol 53: 8649-8663. [ Ref ]
Xu J, Bravo AG, Lagerkvist A, Bertilsson S, Sjoblom R, et al. (2015) Sources and remediation techniques for mercury contaminated soil. Environ Int 74C: 42-53. [ Ref ]
Shu R, Dang F, Zhong H (2016) Effects of incorporating differently treated rice straw on phyto availability of methylmercury in soil. Chemosphere 145: 457-463. [ Ref ]
Eckley CS, Blanchar P, McLennan D, Mintz R, Sekela M (2015) Soil-air mercury flux near a large industrial emission source before and after closure (Flin Flon, Manitoba, Canada). Environ Sci Technol 49: 9750- 9757. [ Ref ]
Kong H, He J, Gao Y, Wu H, Zhu X (2011) Cosorption of phenanthrene and mercury (II) from aqueous solution by soybean stalk-based biochar. J Agric Food Chem 59: 12116-12123. [ Ref ]
Shen B, Tian L, Li F, Zhang X, Xu H, et al. (2017) Elemental mercury removal by the modified biochar from waste tea. Fuel1 87: 189-196. [ Ref ]
Liebert CA, Hall RM, Summers AO (1999) Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev 63: 507-522. [ Ref ]
Boyd ES, Barkay T (2012) The mercury resistance operon: From an origin in a geothermal environment to an efficient detoxification machine. Front Microbiol 3: 349. [ Ref ]
Lu X, Gu W, Zhao L, Farhan Ul, Haque M, et al. (2017) Methylmercury uptake and degradation by methanotrophs. Sci Adv 3: e1700041. [ Ref ]
Zhou XQ, Hao YY, Gu B, Feng J, Liu YR, et al. (2020) Microbial Communities Associated with Methylmercury Degradation in Paddy Soils. Environ Sci Technol 54: 7952-7960. [ Ref ]
Schaefer JK, Letowski J, Barkay T (2002) mer -mediated resistance and volatilization of Hg (II) under anaerobic conditions. Geomicrobiol J 19: 7-102. [ Ref ]
Schaefer JK, Yagi J, Reinfelder JR, Cardona T, Ellickson KM, et al. (2004) Role of the bacterial organomercury lyase (MerB) in controlling methylmercury accumulation in mercurycontaminated natural waters. Environ Sci Technol 38: 4304-4311. [ Ref ]
Sanz-Sáez I, Pereira-García, Bravo AG, Trujillo L, Pla I Ferriol M, et al. (2022) Prevalence of Heterotrophic Methylmercury detoxifying bacteria across oceanic regions. Environ Sci Technol 56: 3452-3461. [ Ref ]
Cabral L, Giovanella P, Gianello C, Bento FM, Andreazza Ret al. (2013) Isolation and characterization of bacteria from mercury contaminated sites in Rio Grande do Sul, Brazil, and assessment of methylmercury removal capability of a Pseudomonas putida V1 strain. Biodegradation 24: 319-331. [ Ref ]
Wagner-Döbler I (2013) Current research for bioremediation of mercury. Bioremediation of mercury: Current research and industrial applications. Caister Academic Press, Norfolk. [ Ref ]
Brim H, McFarlan SC, Frederickson JK, Minton KW, Zhai M, et al. (2000) Engineering Deinococcus radiourans for metal remediation in radioactive mixed waste environments. Nat Biotechnol 18: 85-90. [ Ref ]
Bae W, Mehra RK, Mulchandani A, Chen W (2001) Genetic engineering of Escherichia coli for enhanced uptake and bioaccumulation of mercury. Appl Environ Microbiol 67: 5335-5338. [ Ref ]
Bae W, Wu CH, Kostal J, Mulchandani A, Chen W (2003) Enhanced mercury biosorption by bacterial cells with surface-displayed MerR. Appl Environ Microbiol 69: 3176-3180. [ Ref ]
Ruiz ON, Alvarez D, Gonzalez-Ruiz G, Torres C (2011) Characterization of mercury bioremediation by transgenic bacteria expressing metallothionein and polyphosphate kinase. BMC Biotechnol 11: 82. [ Ref ]
Chen S, Wilson DB (1997) Construction and characterization of Escherichia coli genetically engineered for bioremediation of Hg (2+)-contaminated environments. Appl Environ Microbiol 63: 2442- 2445. [ Ref ]
Vigneron A, Cruaud P, Aubé J, Guyoneaud R, Goñi-Urriza M (2021) Transcriptomic evidence for versatile metabolic activities of mercury cycling microorganisms in brackish microbial mats. NPJ Biofilms Microbiomes 7: 83. [ Ref ]