Journal Name: Journal of Applied Microbiological Research
Article Type: Research
Received date: 13 September, 2017
Accepted date: 09 October, 2017
Published date: 18 October, 2017
Citation: Hwanhlem N, Suwansri S, Domig KJ, Saimmai A (2017) Study of Bacterial Diversity by PCR-DGGE Technique and Screening of Bacteriocin-Producing Lactic Acid Bacteria in Feces of Pig Cultured by Traditional Method in Southern Thailand. J. Appl. Microb. Res. Vol: 1, Issu: 1 (01-06).
Copyright: © 2017 Hwanhlem N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
The bacterial diversity of 15 feces of pig cultured by traditional method in southern Thailand were collected and investigated by using both culture-dependent and culture-independent method—polymerase chain reaction and denaturing gradient gel electrophoresis (PCR–DGGE). Six genera including Brevibacterium, Carnobacterium, Enterococcus, Lactobacillus, Lactococcus and Leuconostoc were observed in pig feces by culture-dependent method. Whereas eight genera including Bifidobacterium, Campylobacter, Clostridium, Enterococcus, Lactobacillus, Porphyromonas, Prevotella and Staphylococcus were observed by analyzing the DNA directly extracted from pig feces. Neutralized Cell Free Supernatants (NCFS) of 219 bacteria isolated from pig feces were also screened for bacteriocin-producing lactic acid bacteria. Among 219 isolates, no any isolate showed an inhibition zone against indicator strains when tested by agar well diffusion assay. This indicated that no bacteriocin-producing strains were found from bacteria isolated from feces of pig.
Keywords
Bacterial diversity; PCR-DGGE; Pig feces; Bacteriocin; Lactic acid bacteria.
Abstract
The bacterial diversity of 15 feces of pig cultured by traditional method in southern Thailand were collected and investigated by using both culture-dependent and culture-independent method—polymerase chain reaction and denaturing gradient gel electrophoresis (PCR–DGGE). Six genera including Brevibacterium, Carnobacterium, Enterococcus, Lactobacillus, Lactococcus and Leuconostoc were observed in pig feces by culture-dependent method. Whereas eight genera including Bifidobacterium, Campylobacter, Clostridium, Enterococcus, Lactobacillus, Porphyromonas, Prevotella and Staphylococcus were observed by analyzing the DNA directly extracted from pig feces. Neutralized Cell Free Supernatants (NCFS) of 219 bacteria isolated from pig feces were also screened for bacteriocin-producing lactic acid bacteria. Among 219 isolates, no any isolate showed an inhibition zone against indicator strains when tested by agar well diffusion assay. This indicated that no bacteriocin-producing strains were found from bacteria isolated from feces of pig.
Keywords
Bacterial diversity; PCR-DGGE; Pig feces; Bacteriocin; Lactic acid bacteria.
Introduction
During last 10 years, bacterial diversity from several sources have been studied such as soil [1], Philippine fermented food products [2], traditional spontaneously fermented Lambic beer [3], human intestinal microbial flora [4], porcine gastrointestinal ecosystem during weaning transition [5] and gastrointestinal tracts of food animal species [6], etc. Most studies of bacterial diversity in several sources use a culturing method that has limitations such as time consuming, limited in terms of both discriminating ability and accuracy and revealing. Some bacterial diversity has been underestimated because the majority of studies were based on methods for culturing organisms, which can characterize only a small fraction of all bacteria living in those sources due to the fact that a large proportion of bacteria are not culturable [7]. Recently, several molecular techniques based on total community DNA extracted from several sources have been widely applied for assessing the microbial diversity [8-10]. The Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis (PCR-DGGE) is one the molecular techniques which has been used to monitor bacterial diversity and community structure due to reliable, reproducible, rapid and greater detection and identification potential than culture based methods [11-13].
Studies of the gut microbiota in pig have been performed by using both culture-dependent techniques and several molecular tools and each of those methods shows its advantage to measure a different aspect of the gut microbial community [14]. Typically, the culture-dependent techniques can sample a minor portion of the bacteria which may not be detected by molecular methods. Compared to a cloning approach, DGGE has the advantage of being less time-consuming and expensive, and has especially great potential for the straight forward comparison of the bacterial community structures from different samples [15]. The gut bacterial diversity with single molecular method may have been underestimated since individual bacterial taxa present in smaller number will not be detected owing to PCR bias. Study of microbial biodiversity in pig feces by using PCRDGGE techniques may help in isolating and identifying either new or potential microorganisms for various applications in human and animal such a bacteriocin producing lactic acid bacteria (LAB) [16,17].
LAB are Gram-positive, non-spore forming cocci, coccobacilli or rods. They have generally anaerobic respiration and lack catalase [16,18]. Based on sugar fermentation patterns, two broad metabolic categories of LAB exist: homofermentative and heterofermentative. During fermentation these bacteria do not produce only lactic acid but they are also known to produce and excrete compounds with antimicrobial activity such a bacteriocins [16,19].
Bacteriocins of LAB are antimicrobial substances that are ribosomally synthesized, releasing bioactive peptides or peptide complexes with bactericidal or bacteriostatic effects. They are secondary metabolite products secreted to inhibit the growth of similar and/or competitive bacterial strains. Most of bacteriocins are small, basic (a net positive charge at neutral or slightly acidic pH) and amphiphilic in nature. They vary in spectrum and mode of activities. They also display different molecular structures, molecular masses, thermostabilities, pH ranges of activities and genetic determinants [17,20]. Currently, bacteriocin producing LAB are extensively studied due to their Generally Recognized As Safe (GRAS) status, since they prevent the growth of many pathogenic and spoilage bacteria such as Staphylococcus aureus, Listeria monocytogenes, Escherichia coli, Pseudomonas spp., Bacillus spp. and Clostridium spp. [17].
Consequently, the objectives of this study were to investigate bacterial diversity by using both the culturedependent method and the culture-independent method of PCR-DGGE and to screen bacteriocin-producing LAB from feces of pig cultured by traditional method in southern Thailand, which might be used as potential information in further research.
Materials and Methods
Sample collection and isolation of bacteria from pig feces
Fifteen fecal samples were collected from pig farm located in southern Thailand where cultured by traditional methodno any antibiotics were used neither in feed nor therapeutic purposes. Samples (25 g) were aseptically transferred to 225 mL of physiological saline solution (0.85% NaCl, 0.1% peptone) to obtain a 1:10 dilution and then shaken for 1 min. Appropriate decimal dilutions (10−4 to 10−6) were prepared in sterile physiological saline solution and spread on de Man, Rogosa and Sharpe (MRS, Hi-Media, India) agar plate. Plates were then incubated at 37ºC for 24 h in anaerobic condition. Morphologically distinct colonies were selected randomly, individually picked and streaked on MRS agar. This procedure was repeated in order to purify the isolates. The isolates were tested for catalase by placing a drop of 3% hydrogen peroxide solution on the cells. Immediate formation of bubbles indicated the presence of catalase in the cells. The isolates were Gram stained and observed under light microscopic [21]. Bacterial isolates were maintained at -20ºC in MRS broths containing 30% glycerol (Scharlab, S.L., USA). For all experiments, the strain was sub cultured twice at 37°C in MRS broth for 24 h.
Indicator bacterial strains and growth conditions
All indicator strains were obtained from Oniris (Ecole Nationale Nantes Atlantique Vétérinaire, Agroalimentaire et de l’Alimentation, Nantes, France) and INRA (Institut National de la Recherche Agronomique, Nantes, France). Growth media, growth condition and source of indicator strains are shown in Table 1. Strains were maintained as frozen stocks at −80 °C in a cryoprotective medium containing 30% glycerol. For all the experiments, strains were sub cultured twice in each growth medium and growth condition which specific to each strain.
Table1: Growth media, growth condition and source of indicator strains.
| Indicator strains | Sources | Growth medium | Growth conditions (°C/h) |
|---|---|---|---|
| Brochothrix thermosphacta DSMZ 20171T | Oniris | BHI | 25 °C/ 24 h |
| Carnobacterium maltaromaticum NCDO 2762 | Oniris | BHI | 30 °C/ 24 h |
| Escherichia coli CIP 76.24 | Oniris | BHI | 37 °C/ 24 h |
| Listeria innocua CIP 80.11T | Oniris | BHI | 30 °C/ 24 h |
| Listeria ivanovii DSMZ 20750T | INRA | BHI | 30 °C/ 24 h |
| Lactobacillus sakei subsp. sakei | INRA | MRS | 37 °C/ 24 h |
| Pediococcus pentosaceus DMST 18752 | INRA | MRS | 37 °C/ 24 h |
| Staphylococcus aureus CIP 76.25 | Oniris | BHI | 37 °C/ 24 h |
Screening of bacteriocin-producing LAB
Preparation of cell free supernatants (CFS) and neutralized CFS (NCFS):The isolates (described in section 4.1) were grown in MRS broth at 37 ºC for 24 h. CFS was obtained by centrifugation (9500 g for 10 min at 4 ºC). NCFS was prepared by adjusting the pH to 7.0 by means of 6 N NaOH to exclude the antimicrobial effect of organic acids. Inhibitory activity from hydrogen peroxide (H2O2) was eliminated by the addition of 1 mg/mL catalase. Samples were heated at 100 ºC for 10 min to inhibit enzyme activity [16,21].
Determination of bacteriocin-producing strain by agar well diffusion assay:BHI or MRS soft agar (1% agar, w/v) was seeded with 106 colonies forming units (CFU) per mL of indicator bacterial strains Table 1, mixed and poured into sterile Petri dishes. After setting, agar wells of 5 mm in diameter were made by using a sterile cork borer. Aliquots (50 μL) of NCFS prepared from section 4.3.1 were placed in wells. Plates were incubated overnight at the optimum temperature of each indicator strain as shown in Table 1, the inhibition zones were then observed and recorded [17,22].
Identification of the selected bacteria
Total DNA was extracted from overnight culture of the 27 selected bacteria (from section 4.1, selected randomly) by using the peqGOLD Bacterial DNA Kit (VWR International GmbH, Germany) according to the manufacturer recommendations and stored at –20 °C until use. Total DNA was used as template for amplification of the 16S rDNA gene by PCR. The sequencing of the amplified fragments was carried out by Eurofins Genomics AT GmbH (Ebersberg, Germany). The obtained sequences were compared with those available in GenBank database, using the Basic Local Alignment Search Tool (BLAST) at the National Center of Biotechnology Information website http://www.ncbi.nlm. nih.gov.
Genomic DNA Extraction from pig feces and PCR-DGGE Analysis
DNA extraction:Total DNA was extracted from 1 g of pig feces by using Genome DNA Extraction Kit (Tiangen Inc., Beijing) according to the manufacturer’s instructions and stored at −20 °C until use.
PCR amplification of V3 region of 16S rDNA:PCR was performed in a total reaction volume of 50 μL containing EmeraldAmp®GT PCR Master Mix (TAKARA BIO INC., Japan) 25 μL, each primer 2 μL (10 μM), DNA template 2 μL and RNase-free water 19 μL. The primer set, 357F-GC (5′-GC-clamp-CCTACGGGAGGCAGCAG-3′) and 518R (5′-ATTACCGCGGCTGCTGG-3′), spanning the V3 regions of the 16S rDNA was used. A GC-clamp (CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG) was added to the primer 357F for DGGE analysis [15]. The PCR products were generated using an initial denaturation step of 5 min at 94°C. This was followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and elongation at 72°C for 45 s. Then final chain elongation was done at 72°C for 7 min.
DGGE Analysis:PCR products were separated by DGGE system (Bio-Rad Laboratories Ltd., England) according to manufacturer’s instructions. Samples (25 μl of each) were loaded onto 1-mm-thick 8 % (w/v) polyacrylamide gels containing acrylamide: bisacrylamide 37.5:1 and a denaturing gradient of 30–65 % (100 % denaturant corresponds to 7 M urea and 40 % (w/v) formamide). The electrophoresis was conducted with a constant voltage of 35 V for 10 min and 85 V for 16 h at 60°C. Gels were stained with SYBR Gold solution (Thermo Fisher Scientific Inc., Australia) for 15 min, then rinsed three times in sterile water and viewed under UV transillumination. The gel images were photographed using the Gel Documentation system (UVitec Cambridge, England).
Excision and sequencing of the DGGE fragments:The DGGE bands were excised with a sterile scalpel and the DNA of each band eluted in 20 μL of sterile milli-Q water, overnight at 4°C. Five microliters of the eluted DNA from each DGGE band was re-amplified using the conditions as described above. Primer 357F without incorporation of a GC-clamp was used. For sequencing analysis, PCR products were purified with the PCR purification kit (QIAGEN Inc., USA) and used as templates in the sequencing reactions. The samples were analyzed with an automated DNA sequencer. The obtained sequences were compared with those available in GenBank database, using the BLAST at the National Center of Biotechnology Information website.
Results and Discussion
Screening of bacteriocin-producing LAB
In order to screen bacteriocin-producing LAB, two hundred nineteen Gram positive bacteria isolated from the feces of pig cultured by traditional method were tested by agar well diffusion assay against indicator bacterial strains Table 1. Neutralized Cell Free Supernatants (NCFS) obtained from these 219 bacteria did not exhibit the inhibition zones. This indicated that among these isolates, there was no any isolate produce bacteriocin.
Identification of the selected bacteria
Morphology and identification of the 27 selected bacteria are shown in Table 2. When the obtained sequences were compared with those available in GenBank database, it could be divided these 27 isolates into 6 genera including Brevibacterium, Carnobacterium, Enterococcus, Lactobacillus, Lactococcus and Leuconostoc wherewith 11 species including Brevibacterium avium, Brevibacterium casei, Carnobacterium jeotgali, Carnobacterium mobile, Carnobacterium viridans, Enterococcus faecalis, Enterococcus faecium, Lactobacillus plantarum, Lactococcus lactis, Leuconostoc carnosum and Leuconostoc gelidum. This result indicated that in feces of pig cultured by traditional method in southern Thailand contained a diverse spectrum of bacteria. However, DGGE analysis did not detect most of these strains as DGGE profile result below. This was probably because using MRS agar as some basal medium isolating bacteria from the pig feces. Basically, MRS is a selective medium for lactobacilli but some growth of leuconostocs and pediococci may occur [23].
Table2: Morphological properties and identification of bacteria isolated from feces of pig cultured by traditional method in southern Thailand.
| Isolates | Catalase Test | Gram stain | Shape | Strains | GenBank accession numbers a |
|---|---|---|---|---|---|
| AO1 | - | + | rod | Lactobacillus plantarum | KJ026699 |
| AO2 | + | + | rod | Brevibacterium avium | Y17962 |
| AO3 | - | + | cocci | Leuconostoc carnosum | NR119221 |
| AO4 | - | + | rod | Carnobacterium mobile | NR040926 |
| AO5 | - | + | cocci | Enterococcus faecalis | LT745973 |
| AO6 | + | + | rod | Brevibacterium avium | JX154087 |
| AO7 | - | + | rod | Lactobacillus plantarum | KY038178 |
| KT1 | - | + | cocci | Enterococcus faecalis | EU887827 |
| KT2 | - | + | rod | Carnobacterium jeotgali | NR116460 |
| KT3 | - | + | cocci | Enterococcus faecalis | DQ983196 |
| KT4 | - | + | rod | Carnobacterium jeotgali | LC258159 |
| KT5 | - | + | rod | Carnobacterium viridans | KU179373 |
| KT6 | - | + | cocci | Leuconostoc gelidum | AB004661 |
| KT7 | - | + | cocci | Leuconostoc gelidum | KR857408 |
| KT8 | - | + | cocci | Lactococcus lactis | AB100796 |
| PK1 | - | + | cocci | Lactococcus lactis | FJ429979 |
| PK2 | - | + | cocci | Leuconostoc carnosum | LC096219 |
| PK3 | - | + | cocci | Leuconostoc carnosum | NR040811 |
| PK4 | + | + | rod | Brevibacterium casei | EU937752 |
| PK5 | - | + | cocci | Enterococcus faecium | EU887814 |
| PK6 | - | + | rod | Carnobacterium mobile | X54271 |
| PK8 | + | + | rod | Brevibacterium casei | KT951720 |
| PK9 | - | + | cocci | Enterococcus faecalis | HM776211 |
| TL1 | - | + | cocci | Leuconostoc gelidum | LC279611 |
| TL3 | - | + | rod | Lactobacillus plantarum | NR115605 |
| TL6 | - | + | rod | Carnobacterium viridans | AF425608 |
| TL7 | - | + | rod | Carnobacterium viridans | NR025197 |
| aAccession number of the sequence of the closest relative species identified using the Blast software. | |||||
Bacterial diversity in pig feces by PCR-DGGE analysis
The results from the direct analysis by PCR-DGGE of the bacterial diversity of 15 samples of pig feces collected from pig farms located in southern Thailand were obtained by amplifying the V3 regions of the 16S rDNA gene using primer 357F-GC and 518R. There was high diversity of bacteria in feces of pig cultured by traditional method. On the basis of the DGGE, different bands in the community structure of bacteria were found in each sample. Samples of an extracted DNA from pig feces showed variation in banding patterns when analyzed by PCR-DGGE. The DGGE bands of different bacterial species were separated at different positions in the polyacrylamide gel because different bacterial species have differences in base pair composition [24]. In all patterns, 5-15 bands of various intensities were detected per sample, with 17 bands shared among all samples because they have differences in base pair composition within the variable regions of the 16S rDNA.
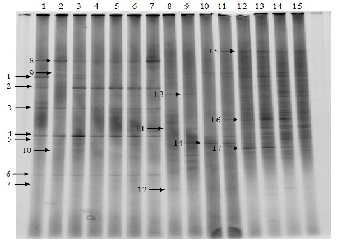
This DGGE profile indicated that in feces of pig cultured by traditional method consist of at least 17 species of bacteria as shown in the (Figure 1). When the 16S rDNA partial sequences from the DGGE bands were compared with the database in GenBank, Band No. 1 and 2 were characteristic of Enterococcus faecalis and Enterococcus faecium, respectively. Band No. 3 and 4 were found to correspond to the sequence of Lactobacillus reuteri and Lactobacillus amylovorus, respectively. Whereas, the other bands were not characteristic of LAB. Band No. 5 and 6 showed 100% similarity to Clostridium coccoides and Clostridium leptum, respectively. Band No. 7 showed 100% similarity to Prevotella albensis. Band No. 8 and 9 were characteristic of Campylobacter avium and Campylobacter fetus subsp. fetus, respectively. Band No. 10, 11 and 12 were characteristic of Porphyromonas cansulci, Porphyromonas gingivalis and Porphyromonas asaccharolytica, respectively. Band No. 13 and 14 showed 100% similarity to Bifidobacterium longum and Bifidobacterium animalis, respectively. The last 3 bands (No. 15-17) were characteristic of Staphylococcus vitulinus, Staphylococcus delphini and Staphylococcus simulans, respectively Table 3.
Figure 1: Denaturing gradient gel electrophoresis (DGGE) profiles of DNA amplicons obtained directly from pig feces. Sequence of bands (1-17) was searched in the GenBank with the BLAST program to determine the closest known relatives of the partial 16S rDNA sequences obtained (Table 3).
Table 3: Closest match identification of DGGE band of pig feces based on 16S rDNA gene sequences in GenBank
| DGGE band no. | Identification from closest match in GenBank | GenBank accession numbers a | % Identity to closest match |
|---|---|---|---|
| Band 1 | Enterococcus faecalis | EU887827 | 100 |
| Band 2 | Enterococcus faecium | AF003921 | 100 |
| Band 3 | Lactobacillus reuteri | AF429625 | 100 |
| Band 4 | Lactobacillus amylovorus | LC064891 | 100 |
| Band 5 | Clostridium coccoides | EF025906 | 100 |
| Band 6 | Clostridium leptum | AF262239 | 100 |
| Band 7 | Prevotella albensis | AJ01168 | 100 |
| Band 8 | Campylobacter avium | NR118510 | 100 |
| Band 9 | Campylobacter fetus subsp. fetus | AF482990 | 100 |
| Band 10 | Porphyromonas cansulci | NR113081 | 100 |
| Band 11 | Porphyromonas gingivalis | KT222964 | 100 |
| Band 12 | Porphyromonas asaccharolytica | L16490 | 100 |
| Band 13 | Bifidobacterium longum | U10152 | 100 |
| Band 14 | Bifidobacterium animalis | AB027536 | 100 |
| Band 15 | Staphylococcus vitulinus | NR024670 | 100 |
| Band 16 | Staphylococcus delphini | NR024666 | 100 |
| Band 17 | Staphylococcus simulans | KP033221 | 100 |
| aAccession number of the sequence of the closest relative species identified using the Blast software. | |||
The results from the direct analysis by PCR-DGGE of pig feces collected from pig farm cultured by traditional method indicated that LAB such as Lact. amylovorus, Lact. reuteri, Ent. faecalis and Ent. faecium were the dominant microorganism because most of the pig fecal samples were found the shared bands in PCR-DGGE profiles. However, some of non-LAB such as Cl. leptum, Por. asaccharolytica, Camp. fetus subsp. fetus and Camp. avium were also the dominant microorganism in feces of pig. Culture-dependent method did not detect most of these strains as the identification of the selected bacteria result above. This was probably because the cell numbers of these strains were higher than those strains for the detection of PCR-DGGE. In addition, biases at the level of DNA extraction and PCR specificity and efficiency could also have been reasons [25].
Our obtained result similar to the report of Peu et al. [26] who studied the dynamics of a pig slurry microbial community during anaerobic storage and management. They reported that several bacterial populations, identified as populations closely related to uncultured Clostridium and Porphyromonas and to Lactobacillus and Streptococcus cultured species commonly isolated from pig feces, remained present and dominant from the rearing build-up to the time of spreading. In similar study, Snell-Castro et al. [27] characterized the microbial diversity in a pig manure storage pit. They reported that the bacterial groups most often represented in terms of phylotype and clone abundance were the Eubacterium, the Clostridium, the Bacillus–Lactobacillus– Streptococcus subdivision, the Mycoplasma and relatives and the Flexibacter–Cytophaga–Bacteroides. The global microbial community structure and phylotype diversity show a close relationship to the pig gastrointestinal tract ecosystem whereas phylotypes from the Acholeplasma–Anaeroplasma and the Clostridium purinolyticum groups appear to be better represented in manure. Archaeal diversity was dominated by three phylotypes clustering with a group of uncultured microorganisms of unknown activity and only distantly related to the Thermoplasmales and relatives.
Conclusion
This study is the first report of the bacterial communities of the feces of pig cultured by traditional method in southern Thailand using culture-dependent and culture-independent (PCR-DGGE) techniques. Our result indicated that in feces of pig cultured by traditional method contained a diverse spectrum of bacteria. Both methods detected different bacterial strains in pig feces. These results indicating that combining and comparing the results obtained from these culture-dependent and culture-independent methods showed the better description of microbial communities of pig feces. Although, we could not obtain any bacteriocin producing LAB from bacteria isolated from these fecal samples as one of our objectives. However, our results provide the important information of bacterial communities of the pig feces for further study.
Acknowledgments
This work was financial supported by National Research Council of Thailand (NRCT) (Project code no. R2559B007). It was also financial supported by Naresuan University, Phitsanulok, Thailand, BOKU University of Natural Resources and Life Sciences, Vienna, Austria and by the Austrian Federal Ministry of Science, Research and Economy (BMWFW) to Dr. Noraphat Hwanhlem within the framework of Ernst Mach- Stipendien, ASEA-UNINET 2017.
1. Garbeva P, van Veen JA, van Elsas JD (2004) Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42: 243–270.
2. Spitaels F, Wieme AD, Janssens M (2014) The Microbial Diversity of Traditional Spontaneously Fermented Lambic Beer. PLoS ONE 9: e95384.
3. Dalmacio LMM, Angeles AKJ, Larcia LLH (2011) Assessment of bacterial diversity in selected Philippine fermented food products through PCRDGGE. Benef Microbes 2: 273–281.
4. Eckburg PB, Bik EM, Bernstein CN (2005) Diversity of the Human Intestinal Microbial Flora. Science 308: 1635–1638.
5. Konstantinov SR, Favier CF, Zhu WY (2004) Microbial diversity studies of the porcine gastrointestinal ecosystem during weaning transition. Anim Res 53: 317–324.
6. Simpson JM, Kocherginskaya SA, Aminov RI (2002) Comparative microbial diversity in the gastrointestinal tracts of food animal species. Integr Comp Biol 42: 327–331.
7. Kozdrój J, Elsas JD van (2000) Application of polymerase chain reactiondenaturing gradient gel electrophoresis for comparison of direct and indirect extraction methods of soil DNA used for microbial community fingerprinting. Biol Fertil Soils 31: 372–378.
8. İnceoǧlu Ö, Hoogwout EF, Hill P, van Elsas JD (2010) Effect of DNA Extraction Method on the Apparent Microbial Diversity of Soil. Appl Environ Microbiol 76: 3378–3382.
9. Tanase AM, Mereuta I, Chiciudean I (2015) Comparison of Total DNA Extraction Methods for Microbial Community Form Polluted Soil. Agric Agric Sci Procedia 6: 616–622.
10. Chen M, Wu B-L, Chen T (2016) The impact of different DNA extraction methods on the analysis of microbial diversity of oral saliva from healthy youths by polymerase chain reaction-denaturing gradient gel electrophoresis. J Dent Sci 11: 54–58.
11. Cocolin L, Alessandria V, Dolci P (2013) Culture independent methods to assess the diversity and dynamics of microbiota during food fermentation. Int J Food Microbiol 167: 29–43.
12. Garofalo C, Bancalari E, Milanović V (2017) Study of the bacterial diversity of foods: PCR-DGGE versus LH-PCR. Int J Food Microbiol 242:24–36.
13. Liu T, Jia T, Chen J (2017) Analysis of microbial diversity in Shenqu with different fermentation times by PCR-DGGE. Braz J Microbiol Publ Braz Soc Microbiol 48: 246–250.
14. Schokker D, Zhang J, Vastenhouw SA (2015) Long-lasting effects of earlylife antibiotic treatment and routine animal handling on gut microbiota composition and immune system in pigs. PloS One 10:e0116523.
15. Li X, Nan X, Wei C, He H (2012) The gut bacteria associated with Camponotus japonicus Mayr with culture-dependent and DGGE methods. Curr Microbiol 65: 610–616.
16. Hwanhlem N, Chobert J-M, H-Kittikun A (2014) Bacteriocin-producing lactic acid bacteria isolated from mangrove forests in southern Thailand as potential bio-control agents in food: Isolation, screening and optimization. Food Control 41: 202–211.
17. Hwanhlem N, Ivanova T, Biscola V (2017a) Bacteriocin producing Enterococcus faecalis isolated from chicken gastrointestinal tract originating from Phitsanulok, Thailand: Isolation, screening, safety evaluation and probiotic properties. Food Control 78: 187–195.
18. Pringsulaka O, Thongngam N, Suwannasai N (2012) Partial characterisation of bacteriocins produced by lactic acid bacteria isolated from Thai fermented meat and fish products. Food Control 23: 547–551.
19. Cizeikiene D, Juodeikiene G, Paskevicius A, Bartkiene E (2013) Antimicrobial activity of lactic acid bacteria against pathogenic and spoilage microorganism isolated from food and their control in wheat bread. Food Control 31: 539–545.
20. H-Kittikun A, Biscola V, El-Ghaish S (2015) Bacteriocin-producing Enterococcus faecalis KT2W2G isolated from mangrove forests in southern Thailand: Purification, characterization and safety evaluation. Food Control 54: 126–134.
21. Schillinger U, Lucke FK (1989) Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol 55: 1901–1906.
22. Hwanhlem N, Ivanova T, Haertlé T (2017b) Inhibition of food-spoilage and foodborne pathogenic bacteria by a nisin Z-producing Lactococcus lactis subsp. lactis KT2W2L. LWT - Food Sci Technol 82: 170–175.
23. De MAN JC, Rogosa M, Sharpe ME (1960) A Medium for the Cultivation of Lactobacilli. J Appl Bacteriol 23: 130–135.
24. Ercolini D (2004) PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J Microbiol Methods 56: 297–314.
25. Madoroba E, Steenkamp ET, Theron J (2011) Diversity and dynamics of bacterial populations during spontaneous sorghum fermentations used to produce ting, a South African food. Syst Appl Microbiol 34: 227–234.
26. Peu P, Brugère H, Pourcher A-M (2006) Dynamics of a Pig Slurry Microbial Community during Anaerobic Storage and Management. Appl Environ Microbiol 72: 3578–3585.
27. Snell-Castro R, Godon J-J, Delgenès J-P, Dabert P (2005) Characterisation of the microbial diversity in a pig manure storage pit using small subunit rDNA sequence analysis. FEMS Microbiol Ecol 52: 229–242.