Journal Name: Journal of Applied Microbiological Research
Article Type: Review
Received date: 27 February, 2023
Accepted date: 13 April, 2023
Published date: 20 April, 2023
Citation: Hernandez M, Bose D (2023) The HOPE Method: Reverse Engineering Antibodies of recovered Patients and Bioproteins. J Appl Microb Res. Vol: 6 Issu: 1 (08-19).
Copyright: 2023 Hernandez M et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
This is a research proposal that describes a method that attempts to use computational models to reverse engineer antibodies of recovered patients without the use of its genes found in effector B cells or the use of memory B cells samples of recovered patients. Most effector B cells are found in bone marrow and not in the serum, thus making it difficult to sample effector B cells from donors. If we concentrate on COVID-19 treatments, even though current development of monoclonal antibodies specific for SARS-CoV-2 has been fortunate to find effector B cells and memory B cells specific for SARS-CoV-2 in the serum, there is a possibility that potent antibodies found in serum whose effector B cells or memory B cells specific for SARS-CoV-2 are not detected in samples of COVID-19 survivors for the development of COVID-19 monoclonal antibodies specific against SARS-CoV-2. Thus, potentially missing an opportunity for the development of potent monoclonal antibodies specific for SARSCoV- 2.
The following is a method, the authors have named the HOPE method, for the development of genetically engineered monoclonal antibodies by studying the neutralizing antibodies (NAb) or broadly neutralizing antibodies (bNAb) of recovered patients of any viral infectious disease. “HOPE” is not an acronym, but named “HOPE” as a symbol of hope specifically for immunocompromised patients that may find more benefit from this proposed treatment.
The HOPE method can also be applied for the development of mAbs specific against non-viral pathogens and tumor neoantigens although efficacy and specificity towards such end targets must be evaluated. The ultimate goal of the HOPE method is to learn from the immune system’s best response to pathogens and tumors and attempt to mimic such response in the development process of mAb. A few steps in the HOPE method can also be used in bio-manufacturing of bioproteins.
Keywords
Neutralizing monoclonal antibodies, Broadly-neutralizing antibodies, COVID-19, Genetically engineered recombinant monoclonal antibodies, precision medicine, Bioproteins.
Abstract
This is a research proposal that describes a method that attempts to use computational models to reverse engineer antibodies of recovered patients without the use of its genes found in effector B cells or the use of memory B cells samples of recovered patients. Most effector B cells are found in bone marrow and not in the serum, thus making it difficult to sample effector B cells from donors. If we concentrate on COVID-19 treatments, even though current development of monoclonal antibodies specific for SARS-CoV-2 has been fortunate to find effector B cells and memory B cells specific for SARS-CoV-2 in the serum, there is a possibility that potent antibodies found in serum whose effector B cells or memory B cells specific for SARS-CoV-2 are not detected in samples of COVID-19 survivors for the development of COVID-19 monoclonal antibodies specific against SARS-CoV-2. Thus, potentially missing an opportunity for the development of potent monoclonal antibodies specific for SARSCoV- 2.
The following is a method, the authors have named the HOPE method, for the development of genetically engineered monoclonal antibodies by studying the neutralizing antibodies (NAb) or broadly neutralizing antibodies (bNAb) of recovered patients of any viral infectious disease. “HOPE” is not an acronym, but named “HOPE” as a symbol of hope specifically for immunocompromised patients that may find more benefit from this proposed treatment.
The HOPE method can also be applied for the development of mAbs specific against non-viral pathogens and tumor neoantigens although efficacy and specificity towards such end targets must be evaluated. The ultimate goal of the HOPE method is to learn from the immune system’s best response to pathogens and tumors and attempt to mimic such response in the development process of mAb. A few steps in the HOPE method can also be used in bio-manufacturing of bioproteins.
Keywords
Neutralizing monoclonal antibodies, Broadly-neutralizing antibodies, COVID-19, Genetically engineered recombinant monoclonal antibodies, precision medicine, Bioproteins.
Introduction
For years different methods to create genetically engineered monoclonal antibodies have been attempted in animal models but found to be too expensive and time consuming for mass production [1].
The following is an attempt to lower the cost of genetically engineered monoclonal antibodies by suggesting a few innovative experiments inspired by published research papers like Barderas R, Benito-Peña E. The 2018 Nobel Prize in Chemistry: phage display of peptides and antibodies [2]. “Neutralizing antibodies (Nab) are antibodies that not only bind to the antigen’s epitope, but at the same time blocks the virus entry to host cells” [3]. Broadly neutralizing antibodies (bNab) are a subtype of neutralizing antibodies that “universally function by targeting epitopes that are highly conserved and exposed on the surface proteins of the variable virus” [4]. The efficacy of Nab have been studied extensively, for example, HIV broadly neutralizing antibodies were found to be the most efficient antibodies in animal models [5]. Upon studying the efficacy of monoclonal antibodies specific against SARS-CoV-2, the importance of the FC region and the FAB region of the mAb became relevant in the determination of the mAb efficacy [6].
Given the efficacy of monoclonal antibodies specific against SARS-CoV-2 during the pandemic, one can hypothesize that neutralizing antibodies and broadly neutralizing antibodies of recovered COVID-19 patients vary in potency and efficacy based on the antibodies ability to most tightly bind their FAB region to their corresponding epitopes as well as its ability of having an efficient FC region. Thus, selecting the neutralizing antibody or broadly neutralizing antibody with these criteria can be used as good guides to try to reverse engineer for the development of potent monoclonal antibodies specific against SARS-CoV-2. One can hypothesize as well that such method in monoclonal antibody production can also be applied in various diseases that produce an adaptive immune response whose antibodies can be used as guides for monoclonal antibody production. We can also hypothesize that analyzing the epitopes that bind to selected neutralizing antibodies and broadly neutralizing antibodies of recovered patients can assist in identifying potential targets, which vaccine development can be directed to, that is by analyzing the epitope’s mRNA sequence that can be added to mRNA vaccine development.
In viral infectious diseases, the neutralizing antibodies are an important part of the HOPE method by selecting the best neutralizing antibody within a population of region, whose FAB component are the most specific to the epitope of the antigen in other words the neutralizing antibodies that binds most compactly to its epitope. Detailing the HOPE method of reverse engineering an antibody of a recovered patient of viral infections even further in particular, the HOPE method is performed with the help of mass spectrometer and cryogenic electron microscope (cryo-EM) to obtain 3D protein models of the neutralizing antibodies (NAb) and run de novo peptide sequencing. Mass spectrometry and computational models are used to decode the linear amino acid sequence. The 3D protein models obtained with cryo- EM may help perfect these computational models with image datasets identifying the amino acids within the protein folded structure and its comparison with the analysis of the mass spectrometer. Computational models can be used to reverse the central dogma by predicting the codon sequence from the amino acid sequence and subsequently, the codon is decoded by another computational model or machine learning algorithm to help predict the mRNA sequence. Computational models to decode the RNA codon from the amino acid sequence can be trained with codon chart analysis. These steps would be done for both the FAB region of an effective antibody against a neoantigen of a tumor or epitope of a pathogen (like SARS-CoV-2 for example, from recovered COVID-19 patients) and the Fc region of a fully human monoclonal antibody that has proven to be effective in prior studies. This is followed by uniting the two mRNA sequences to form the mRNA of a full monoclonal antibody specific to the tumor or pathogen, like SARS-CoV-2. The predicted mRNA sequence of the full monoclonal antibody can be genetically engineered into plasmids and reproduced in yeast cultures with recombinant DNA technology or other cost-effective methods for mass production, as detailed in this paper.
The HOPE method can also develop genetically engineered monoclonal antibodies that is most specific or most tightly bound to its epitopes of antigens of non-viral pathogens from serum samples of recovered patients of nonviral infectious diseases. Such monoclonal antibodies have not been applied clinically and must be evaluated further to determine its efficacy on non-viral infectious diseases. The HOPE method can also be applied to antibodies of oncology patients specific against tumor neoantigens for the development of personalized precision medicine and diagnostic tests. If we venture out further, certain steps of the HOPE method may also potentially be used in material science for mass production of bioproteins whose genes are unknown.
The authors would like to keep HOPE Monoclonal Antibodies (HOPE-mAb) as the nomenclature of the genetically engineered recombinant monoclonal antibodies produced via the HOPE method. Although, subsequent steps around the development of HOPE-mAb may appear specific to COVID-19, the overall methodology can be broadly applied for other diseases or tumors that produce antibodies in recovered patients. HOPE mAbs specific against SARSCoV- 2 can be commercialized more rapidly for in vitro rapid diagnostic COVID-19 tests and for laboratory research use in COVID-19 studies. Rapid tests development and laboratory research use of HOPE mAbs for other diseases may also be possible with HOPE method upon showing its efficacy in binding to their intended epitopes.
Antibodies or Immunoglobulins
Antibodies can also be called immunoglobulins (Ig), [Ig are glycoproteins which are proteins with carbohydrates attached to its peptides]. Antibodies are produced by effector B cells also called plasma cells, which are differentiated B cells or specialized B cells of the humoral immunity. “In addition to the spleen and lymph nodes, memory B cells are found in the bone marrow, Peyer’s patches, gingiva, mucosal epithelium of tonsils, the lamina propria of the gastro-intestinal tract, and in the [blood] circulation” [7]. The basic functional unit of each antibody is an immunoglobulin (Ig) monomer consisting of 2 heavy chains and 2 light chains [8]. The antibody variation also called isotypes and they vary based on the Ig monomer’s type of heavy chains, which differ in size and are named by Greek letters: α, δ, ε, γ and μ [9]. There are two types of light chains variations, lambda (λ) and kappa (κ), which vary slightly between antibody isotypes, thus will not be mentioned while describing the different antibody isotypes. “Genes encoding antibody heavy and light chains were generated by rearranging different gene segments [V and J (joining) for light chains and V, D (diversity), and J for heavy chains” [10]. Antibodies that are commonly found as only one immunoglobulin monomer are IgG, IgD, and IgE antibody isotypes. The IgG monomer contains 2 gamma (γ) heavy chains and has a prolonged half-life in comparison to other antibodies, thus are usually detectable for longer periods of time. The IgD monomer contains 2 delta (δ) heavy chains. The IgE monomer contains 2 epsilon (ε) heavy chains. Ig that are dimeric in other words that are commonly united as a pair of 2 immunoglobulin monomers. Examples of dimeric Ig are the IgA antibodies, which contains 2 IgA monomers united and each with 2 alpha (α) heavy chains. It is interesting to note that IgA antibodies can be present in 2 forms; mainly dimeric in secretory tissue and occasionally monomeric in the serum. Antibodies isotypes can also be tetrameric (4 immunoglobulin monomers) and pentameric (5 immunoglobulin monomers). IgM also has two forms depending on whether it is membrane bound to B cells or unbound. The membrane bound IgM is a monomer and is the first membrane bound antibody during the development of all B cells, but as the B cells differentiate and mature their antibody isotype changes. The unbound IgM tends to unite with other IgM to predominantly form pentameric (5 IgM monomers each with mu (μ) heavy chains), although it can also be seen as tetrameric (4 IgM monomers) in occasions. “During early development, B cells express only IgM-BCR, while IgD is produced later along with IgM by alternative pre-mRNA splicing at mature B cell stages [11-13]. After encountering an antigen, IgM+IgD+ mature B cells undergo CSR to produce IgG, IgA, or IgE isotypes” [14].
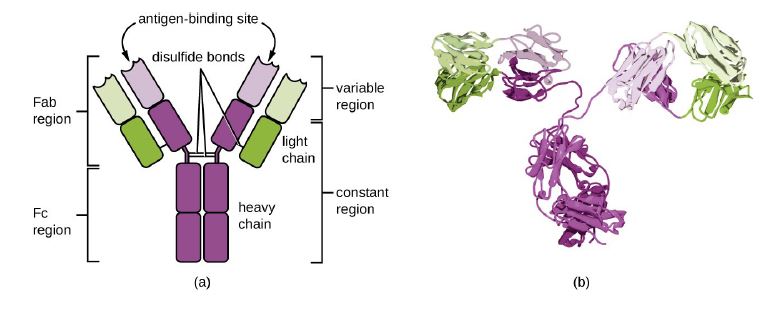
Figure 1:© Parker et al. 2022, Mar 21, 2022 OpenStax. CC BY 4.0 license [16].
A: "The typical four-chain structure of a generic antibody monomer" [17]. The 2 shades of Green indicate the 2 light chains. Each light chain has one variable
(VL) and one constant (CL) domain. The shades of purple indicate the 2 heavy chains. Each Heavy chain has one variable (VH) domain followed by a constant
domain (CH1), a hinge region, and two more constant (CH2 and CH3) domains [16].
B: "Corresponding three-dimensional structure of the antibody IgG. (Credit b: modification of work by Tim Vickers)" [16].
The Ig monomer is a “Y”-shaped molecule as seen in figure 1; containing sites that can bind generally two identical epitopes of the antigen. When antigens are presented to naive B cells by activated T cells in the germinal centers of a lymph node, a series of selection process also called affinity maturation due to variable domain mutations of both heavy and light chain that enhance affinity [15]. During affinity maturity, B cells with low affinity towards the epitopes die off and only B cells with high affinity towards the epitope are allowed to continue to differentiate via isotype switching (DNA recombination of VDJ in the heavy chains genes constant domain), into long lived plasma cells that produce and secrete antibodies and memory B cells. “The affinity of an antibody for an antigenic determinant describes the strength of binding of a single copy of the antigenic determinant to a single antigen-binding site, and it is independent of the number of sites” [9]. Thus, low affinity binds loosely to an epitope while high affinity binds tightly to the epitope. High affinity tends to target more precisely thus have high specificity towards the targeted epitope. Unlike long lived plasma cells, memory B cells cannot produce antibodies, but they can change to do so upon rapid activation after undergoing clonal expansion and differentiate into long lived plasma cells which can then produce antibodies. Each clone of B cells represents replication of identical cells. Antibodies from the same clone of effector B cells all produce antibodies specific against the same epitope of the antigen since they are all identical cells. These antibodies from one clone of effector B cells are called monoclonal antibodies [16,17].
When a group of different clones of effector B cells produces antibodies for different epitopes found in the same antigen, the antibodies are called polyclonal antibodies. Antigens are molecules or pieces of molecules that are recognized as foreign by the immune system, “typically they can be simple molecules, toxins, chemicals, proteins, carbohydrates, lipids or nucleic acids” [18]. Cells of the innate immune system can recognize antigens in any of its forms, but the cells of the adaptive immune system were first thought to have only recognized antigens in the form of peptides for activation of its T cell receptors and B cell receptors in order for antibodies production to occur within effector B cells. In the last decade, scientists have discovered activation of both T cells and B cells by antigens in the form of carbohydrates as well as peptides [19]. When antibodies bind to self-antigens, diseases like autoimmune diseases may occur. However, B cells specific against self-antigens are usually eliminated in the selection process prior to plasma cell differentiation and antibody production in plasma cells. Nevertheless, caution should be exercised to not select an antibody specific against self-antigen in the selection process of the antibody that is selected from recovered patients during step 1 of the method detailed in this paper.
The antigen-binding regions of the antibodies (known as FAB regions) are like lego pieces that can fit perfectly with other lego like pieces called the epitopes of microbes and tumors. As mutations of the pathogen occur these “lego” pieces of the epitope morph, making the binding of prior antibodies towards the same pathogen less effective. Thus, the immune system must make new antibodies high affinity (that bind more tightly) and high specificity to the morphed epitope once more.
Brief introduction to Monoclonal Antibodies (mAbs)
A monoclonal antibody (mAb) is defined as an antibody derived from a single B cell clone and recognizes a unique epitope [20]. The first monoclonal antibodies were generated in a hybridoma in mice in 1975. It took a little more than 10 years to transition from its creation in vitro with the help of animal models to the first licensed mAb for use in humans: muronomab, a purified mouse mAb specific against human CD3, approved in 1986 primarily for kidney transplant rejection prophylaxis. Therapeutic mAb can be classified per its origin, the suffix in its nomenclature refers to this classification: Human [-umab], Humanized [-zumab] from humanized mice, Chimeric [-ximab] from more than one origin, and Murine [-omab] from mice. “While the primary target of all mAbs are the epitopes of pathogens, this binding can cause multiple effects such as disruption of function of the targeted antigen or elimination of cells or pathogens... These effects may result from either the direct binding to target antigen by the antigen-binding fragment (FAB) region or as result of the activation and recruitment of immune cells or serum complement upon binding to the fragment crystallizable (Fc) region of the Ig” [20].
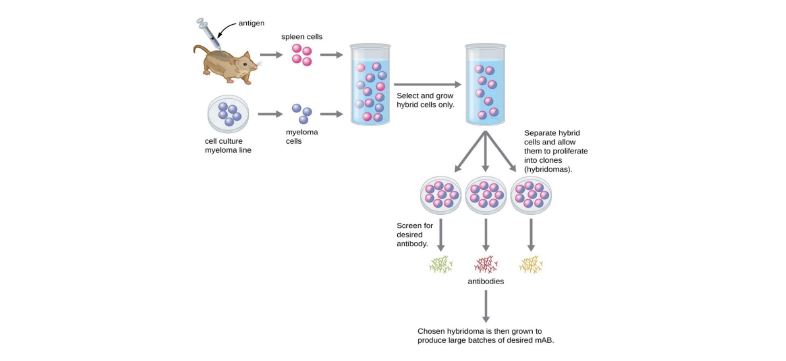
Figure 2:Traditional Monoclonal antibodies (mAbs) production with Hybridoma technology. Either in vivo or in vitro method (not shown in image) used after development of hybridoma tends to be expensive and time consuming. © Parker et al. 2016, Mar 21, 2022 OpenStax. CC BY 4.0 license [17].
The development process of monoclonal antibodies can be divided into traditional mAb production and modern mAb production. The traditional mAb production use hybridoma technology. Modern mAb production use recombinant DNA technology. Hybridoma technology traditionally initiates with the animal models, usually mice, are injected with the antigen which is called the immunization phase. Antigens are complex structures with multiple epitopes and usually provoke a strong immune response, where numerous effector B cells can target the same antigen but each targeting a specific epitope. Remember, effector B cell produce identical antibodies specific for one epitope of the antigen, but since there can be numerous effector B cells, the immune system can produce many different antibodies to the different epitopes in the same antigen. This response is called a polyclonal antibody response, which is also a typical response seen in mice and humans alike. Upon the development of an effective immune response in mice, the effector B cells are isolated in antiserum upon sacrificing the mice and collecting the mice’s spleen. The selected mice effector B cells are then fused with an immortal myeloma cell to form a hybridoma. The hybridoma is now an immortal cell capable of secreting antibodies specific against the antigen that were injected in the mice. Upon developing the hybridoma, manufacturers can either use the in vivo or an in vitro method to mass produce mAb. In vivo method intraperitoneally injects mice with 105 to 110 viable hybridoma cells where mAb can be mass produced within the mice. Weeks later the mAb are harvested from the mice by collecting their ascites fluid. This method requires purification which makes it the less preferred method. In vitro method uses cultures of hybridoma in a laboratory within a special media that allows the hybridoma to mass produce and secrete antibodies within the culture. The antibodies are then collected from the media in a purified form; thus, this method is preferred from the in vivo method [21]. The in vitro method can allow manufacturers to develop monoclonal antibodies, by selecting a single clone of effector B cell specific for one epitope of the antigen. On the contrary the in vivo method where mice with a polyclonal antibody response produce polyclonal antibodies against several epitopes of the same antigen, isolating an antibody in the antiserum for a specific epitope of an antigen is challenging thus this method is usually used for collection of polyclonal antibodies. The use of either method in hybridoma technology is a very expensive and time-consuming process (Fig. 2). It may take weeks of culturing and many liters of media to provide enough mAbs for an experiment or to treat a single patient.
Recombinant antibodies are really recombinant monoclonal antibodies produced by recombinant DNA technology and genetic engineering by using in vitro cloning of animal or human effector B cells. With genetic engineering, the genetic sequence for an antibody’s light and heavy chains are obtained from the effector B cell and inserted into expression vectors, like plasmids, that are then transfected into glycosylation-engineered yeast, bacteria, insect cell lines, transgenic plants or mammalian cells [22,23]. “Given that recombinant antibodies often need to undergo a series of post-translational modifications (such as glycosylation modification), folding, and correct cleavage, antibody drugs with biological activity and low immunogenicity can be produced, therefore, mammalian cells have become the dominant system for the production of recombinant antibodies, especially for full-length monoclonal antibodies” [22]. Recombinant antibodies have been around for the past few decades and mainly used for research and diagnostics applications. It wasn’t until recently that it has been rapidly used for therapeutic applications as well and the market has boomed especially upon the discovery of the Chinese Hamster Ovary (CHO) cell capacity to improve the glycosylation modification that recombinant antibodies need to undergo during antibody mass production. Although genetically engineered lentivirus can also be used to transfect the cell culture media, due to the risk of new viral pandemics, a suggestion from the authors can be to simply breaking up the genetic sequence in various plasmids if the whole sequence cannot fit into a single plasmid to avoid the use of genetically engineered viruses and reduce the risk of accidental or intentional (bio-terrorism) transmission to humans.
COVID19 mAb
Monoclonal antibodies specific for SARS-CoV-2 in early development for COVID-19 that were first developed were “claimed to be fully “human” and were discovered from SARS-CoV-2-immune donors (majority), SARS-CoV immune donors (VIR-7831 and ADG2), immunized humanized immunoglobulin mice (REGN10933 and ABBV47D11), or wild-type mice (ABBV-2B04)” [20]. All of the first mAb specific against SARS-CoV-2 in early development were neutralizing antibodies specific against different epitopes in the spike protein of SARS-CoV-2, specifically against the receptor-binding domain of the S1 subunit.
The SARS-CoV-2 spike (S) glycoprotein is a transmembrane protein and the target of neutralizing antibodies, including all mAbs currently authorized for EUA. The S glycoprotein has two functional subunits that respectively mediate host cell attachment (S1 subunit) and the fusion of the viral and cellular membranes (S2 subunit). Both subunits are essential for the viral entry into the host cell. The S1 subunit is formed by four domains, the most relevant being the N-terminal domain (NTD) and the receptor-binding domain (RBD) [24]. “The major strategy used for rapid isolation of high-efficacy nAbs is reverse transcriptase-polymerase chain reaction (RT-PCR) from single human B cells. In this approach, the SARS-CoV-2 S or RBD protein-specific memory B cells from convalescent or acute-phase COVID-19 patients are sorted by flow cytometry, and single-cell RT-PCR for immunoglobulin [variable region] genes is performed” [25]. Instead of searching for effector B cells which are mainly in bone marrow, the memory B cells found in recovered COVID-19 patient’s serum are collected. These memory B cells can be presented with the antigen, undergo expansion and differentiation into effector B cells within in vitro cultures which can then be transfected into either hybridoma technology (in vivo animal models or in vitro cultures) or recombinant mAb development.
As of 06-03-2022, FDA 2022 Emergency Use Authorization official website, the currently US FDA Emergency Use Authorization (EUA) approved monoclonal antibody specific against SARS-CoV-2 that have not yet obtained full FDA approval for COVID19 are [26]:
• Bebtelovimab (Eli Lilly mAb approved 02-11-2022.)
• EVUSHELD (tixagevimab co-packaged with cilgavimab Astrazeneca mAbs approved 12-08-2021 both mAbs are derived from B cells from COVID-19 survivors.)
The following are prior EUA approved monoclonal antibody specific against SARS-CoV-2 that were discontinued for use in the US due to the Omicron BA.2 sub-variant frequency within the US population and the reduction in efficacy the following mAbs showed during in vitro studies:
• Sotrovimab (GlaxoSmithKline LLC mAb approved 05- 26-2021 developed from a B cell of a SARS-CoV survivor that was stored in 2003 and developed into a broadly neutralizing recombinant engineered mAb in 2021 specifically engineered to extend its half-life. The mAb binds to a conserved epitope that was found in the spike protein of both SARS-CoV and SARS-CoV-2, specifically in the RBD of the S1 subunit.)
• Bamlanivimab and Etesevimab (Eli Lilly mAb approved 02-09-2021 both were developed by screening for B cells in COVID-19 survivors)
• REGEN-COV (Casirivimab and Imdevimab; Regeneron mAb approved 11-21-2020 developed from B cells of transgenic mice and screening for human PBMC cells of COVID-19 survivors)
• Bamlanivimab alone. (Eli-Lilly and AbCellera mAb developed approved 11-09-2020)
The first monoclonal antibody specific against SARSCoV- 2 to obtain US FDA Emergency Use Approval (EUA) was Bamlanivimab alone was developed by both Eli-Lilly and AbCellera by screening B cells specific for SARS-CoV-2, via a high-throughput microfluidic screen, from the first U.S. COVID-19 survivor [25]. Bamlanivimab was followed shortly by REGEN-COV (Casirivimab and Imdevimab; a Regeneron neutralizing mAb approved 11/21/2020), which was shown to be effective enough for EUA upon completion of human clinical trials that started in June 2020. Both Casirivimab and Imdevimab are human immunoglobulin G-1 (IgG1) monoclonal antibody (mAb) produced by recombinant DNA technology in Chinese hamster ovary (CHO) cell suspension culture. “Casirivimab was identified from VelocImmune hAb transgenic mice immunized with a DNA plasmid encoding SARS-CoV-2 S protein, followed by a booster of injected recombinant S protein. Meanwhile, imdevimab was identified from isolated PBMCs of three human donors previously infected with SARS-CoV-2” [25]. Soon other EUA were given to solo or combined human IgG-1 neutralizing mAb specific against SARS-CoV-2, all targeting the spike protein of SARSCoV- 2 and all produced by recombinant DNA technology in Chinese hamster ovary (CHO) upon obtaining samples of the available effector B cells from COVID-19 survivors or humanized transgenic mice. This comparison was done upon comparing the description section of the FDA fact sheet for healthcare providers of each available COVID-19 mAb with EUA available to the public.
Reflection on the Inefficacy of mAbs
Most monoclonal antibody therapy specific against SARS-CoV-2 remained effective until the appearance of the Omicron BA.2 subvariant in December 2021. It can be hypothesized that the affinity of a few of these monoclonal antibody therapies did not withstand fifty or more mutations often found in the Omicron subvariant due to imperfect binding. “Indeed, the broad distribution of bamlanivimab plus etesevimab has been paused in the United States because the Omicron variant has markedly reduced in vitro susceptibility to bamlanivimab and etesevimab; and therefore, this regimen is not expected to provide clinical benefit for patients with Omicron infection” (US department of Health and Human Services 2022) [27]. Similar findings were observed for REGEN-COV and Sotrovimab which were also paused in the US, thus may not be administered for treatment of COVID-19 under the Emergency Use Authorization until further notice by the Agency. Although other countries have not made such restrictions, but due to the frequency of Omicron BA.2 sub-variant in the US such precautions were made. Despite being Sotrovimab a bNab showing efficacy in other Omicron subvariants, data from in vitro studies showed a significant decrease in efficiency against Omicron BA.2 and causing a debate between the GSK scientists and academic scientist. The National Institute of Health 2022 (NIH COVID-19 treatment guidelines on SARS-CoV-2 variants and susceptibility to Anti-SARS-CoV-2 Monoclonal Antibodies) concluded that in vivo studies would be unlikely to be active and paused distribution and application of Sotrovimab for EUA on April 2022 [28].
Immune evading SARS-CoV-2 variants, which may mark the beginning of antigenic drift of SARS-CoV-2, may potentially continue to emerge and co-evolve when herd immunity is reached, with implications for reinfection, vaccines, and both mAb and polyclonal antibody therapeutics. Monitoring resistance of mAbs in circulating new variants will be key to define whether some of the developed mAbs should be discontinued or if different combinations of clinical-stage mAbs should be investigated. Many clinical mAbs are not sensitive (in vitro or in vivo) to the mutations present in the current variants of concern (VOCs). Instead of producing mAbs à la carte, targeting each individual VOCs, focus should go towards continuing targeting highly conserved epitopes of the viral antigen by producing mAbs from broadly neutralizing antibodies specific against SARS-CoV-2. This is a more efficient method that needs to be explored in order to increase efficacy and reduce the cost of the research and development of COVID-19 mAb.
Evolution of the Adaptive Immune System
The adaptive immune system has evolved for millions of years. It evolved from invertebrates to first Jawless Invertebrates, whose antibodies had a decameric-shape structure, to the Jawed vertebrates whose antibodies had a y-shape [29,30]. Did the immune system evolve differently within the vertebrates from decameric shape to y shape to perfect the antigen binding site to fit perfectly to the different antigens each were exposed most frequently to?
The adaptive immune system is amazing in that the antibodies it produces protects the body not only by binding to the pathogens, but it also undergoes an extensive selection process thus only the B-cell, whose antigen receptor best binds perfectly to the pathogen is allowed to proceed to differentiate and make antibodies to be mass produced. Per Alberts, “As predicted by the clonal selection theory, all antibody molecules made by an individual B cell have the same antigen-binding site [consisting of a heavy chain and light chain variable region]. The first antibodies made by a newly formed B cell are not secreted. Instead, they are inserted into the plasma membrane, where they serve as receptors for antigen” [9]. During B cell development, first heavy chain rearrangement and selection occurs followed by light chain rearrangement and selection for the formation of naive B cells not yet bound to antigens. Upon presentation of the antigen to the naive B cell in the germinal centers of lymph nodes, the precise selection process of the binding site specific for the epitopes of the antigen begins [31].
How can we take advantage of the selection process that assures proper binding between antibodies and pathogens, already being performed by a potent immune system of a recovered patient for the development of monoclonal antibodies? Using the genes of antibodies of recovered patients for drug development has proven to work thus far—as that is how the current EUA monoclonal antibodies specific for SARS-CoV-2 were created.
But can we improve further by making the selection process even better and making it more cost effective? Are we limited to only using the genes of antibodies whose effector B cells or specific memory B cells that are collected in the sample of recovered patients?
Would it be possible to reverse engineer antibodies of recovered patients without knowing its gene in the effector B cell that produced it? If we develop a method to do this, can we reverse engineer any biological protein without knowing its gene and apply it in mass producing bioproteins for sustainable material science? The HOPE mAb method provides a step by step process that attempts to address these questions.
Material and Methods
The HOPE method is shown in the following steps, as shown in figure 3.
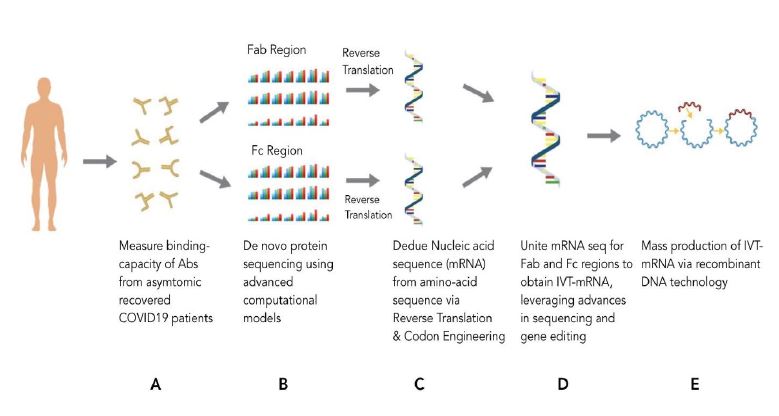
Figure 3:The HOPE method proposed by Michellie Hernandez with the following steps:
A: Studies of antibodies from recovered patient population to select the best antibody to reverse engineer.
B: Reverse engineering Fab and Fc region of antibody to its linear amino acid sequence with de novo protein sequencing and computational models.
C: Reverse engineering Fab and Fc region from its linear amino acid sequence to its codon sequence followed by its mRNA sequence with reverse translation
and codon engineering computational models.
D: Union of the mRNA sequence of both Fab and Fc region with genetic engineering.
E: Creation of in vitro transcribed mRNA (IVT-mRNA) within a plasmid for induction of mass production of HOPE mAb via recombinant DNA technology
in yeast or or larger plant cultures.
Figure by Deb Bose.
A few steps in the method process are subject to US patents. Most of the following method was previously made public by the primary author [32,33].
(1) The first step in the method process will vary depending on whether the desired antibody for reverse engineering is specific against a pathogen, such as SARSCoV- 2, which will require studies of antibodies from recovered patient population or if the antibody for reverse engineering is a natural antibody specific against a tumor. In this case, since neoantigens of tumors are unique to each cancer patient, reverse engineering natural antibody specific against a tumor will require personalized individual study of the cancer patient’s own natural antibodies specific to the neoantigens of the tumor to select the antibody that is most specific (most tightly binds) to the neoantigen. Thus, the HOPE mAb specific to the neoantigen will be a specific therapeutic or diagnostic test for that patient and not for the general public. Per Pinto et al. (2020), confirmed the binding of the antibody to its epitope by cryo-electron microscopy and binding assays in his study, although other approaches to confirm antigen’s specificity might have been improved upon since then [34].
The following is specific to SARS-CoV-2, but may be applied to other viral pathogens that produce an antibody response: Conduct an antibody study in asymptomatic recovered COVID-19 patients to collect serum samples and test for neutralizing antibodies.
Asymptomatic COVID-19 patients tend to be a good indicator for effective neutralizing antibodies, since their immune system created a strong enough response to avoid any COVID-19 symptoms. Since there are no assay to unbind the antibody from the epitope as of yet developed, cryo- Em and spectrometer of each antibody should be done prior to pathogen exposure so images and data analysis can be used in computational models in the following steps. Once antibody analysis is complete, exposure of the pathogen to the antibody is is done and test for neutralizing antibodies. Upon detection of neutralizing antibodies, test the specificity of each sampled neutralizing antibody to bind to SARS-CoV-2 spike protein with mass spectrometer or cryo-Em and binding assays. Essays with nonpathogenic remnant of SARS-CoV-2 containing spike proteins can be used to test for the specificity of the antibodies to the spike protein in order to reduce the costs and need of BSL3 labs. One can confirm with tandem mass spectrometry analysis to select the best neutralizing antibody with the most cross binding between antibody and spike protein and cryo-EM to obtain a 3D-protein model structure of the FAB region of the antibody [35]. Analyze the epitope that binds to the neutralizing antibody with the most specificity and evaluate that epitope’s mutation history. Analyze the epitope that binds to the neutralizing antibody with the most specificity and evaluate that epitope’s mutation history. Select the most effective neutralizing antibody specific against SARSCoV- 2 that binds the most tightly to the epitope and has the least known history of mutations. Ideally a screening test that is capable of binding and unbinding to the epitope in the medium can be created to reduce the cost and time consumption. Otherwise prior to screening test the binding capability to epitope for each of the tested antibodies in Step 1 of this method and 3D protein models will have to be created for all the antibodies. But if an assay unbinding the antibody from the epitope without affecting the FAB region structure is able to be created, only the selected antibody that best binds to epitope with the least history of mutation will have to be studied by mass spectrometry analysis and cryoEM to produce a 3D protein images and data analysis of the antibody.
(2) Using HPLC mass spectrometer and cryogenic electron microscopy (cryo-EM), de novo peptide sequencing, computational models can essentially try to reverse the Central Dogma of biology through a series of algorithms to decode the mRNA sequence of the FAB region (the binding site of the antibody to the antigen) of the selected neutralizing antibody in Step 1 that was most specific antibody against SARS-CoV-2. To do this the mass spectrometry data analysis and cryo-EM images obtained in Step 1 can be used to obtain the 3D model of the antibody. De novo peptide sequencing by deep learning can help predict the linear amino acid sequence from a given protein that has undergone mass spectrometer analysis in computer models [36]. The identification of the amino acids sequences also called the linear protein structure of amino acids of a known protein has been recorded before in 2019 [37]. Both instruments are needed for the computational models since they will be trained previously with cryo-Em images databases that compared the images and data analysis of the mass spectrometer of other antibodies of known genetic sequences to test the models. The authors suggest to train the computational models with database of proteomic images to identify the amino acids in the protein first to help follow the amino acid motion during the unfolding of the protein to its linear form that is suggested by mass spectrometer. Further computational models such as Gene Design Modules, can decode the codon of the amino acid sequence via reverse translation also known as back translation in computational medicine and codon engineering [37-40]. “Reverse Translate accepts a protein sequence and uses a codon usage table to generate a graph that can be used to find regions of minimal degeneracy at the nucleotide level [39].” Thus, this step in the HOPE method will have to be tested to confirm if it improves the prediction rate of current de novo peptide sequencing algorithms. This can take us a step further in the series of computational models with machine learning or deep learning to decode the mRNA sequence of the FAB region of the antibody that must be done. Once the linear amino acid sequence is predicted by de novo peptide sequencing computational models, the use of RNA codon tables to achieve reverse translation also called back translation and codon engineering can be done basically, to decode the amino acid sequence to codon [37-39,41]. The final computational model used will be to predict the mRNA sequence from the codons in which the models can be trained with codon chart analysis of known mRNA and known codons of a given protein.
(3) Obtain the mRNA sequence of the Fc region of a either a human antibody of a COVID-19 survivor or a fully human monoclonal antibody (mumab) that has shown to be effective in animal studies or human organ chip disease modeling research studies [42]. The constant region of the antibody contains the FC region which along with the specificity of the FAB region has been suggested in prior studies to play an important role in the effectiveness of the neutralizing antibody [6]. Follow the same procedures as above: Use HPLC mass spectrometry or cryo-EM to create a 3D model protein structure of the Fc region and machine learning computational models like de novo peptide sequencing, to decode the mRNA sequence from the 3D modeling.
(4) Unite both mRNA sequences obtained in steps 2 and 3 so the union of the two encodes a complete fully human monoclonal antibody (mumab) effective against SARSCoV2. This should be possible with the advances of sequencing technologies in recent years with gene editing done in prior monoclonal antibody generations [43].
(5) Obtain the mRNA sequence of an In Vitro transcribed mRNA (IVT mRNA) encoding the combined mRNA sequence. Per Schlake et al., mRNA prepared by in vitro transcription (IVT) is increasingly appreciated as a drug substance for delivery of recombinant proteins [44].
(6) To make IVT mRNA production more cost-effective, test mass production of IVT mRNA with recombinant DNA technology. Synthesize a synthetic DNA sequence that upon transcription will transcribe the mRNA sequence in Step 5 (IVT mRNA without the delivery system) via reverse transcription and genetic engineering. The synthetic DNA is inserted into a plasmid and with the use of recombinant DNA technology in E.Coli or yeast culture, clones of IVT mRNA could be reproduced [45]. These IVT mRNA will induce translation from mRNA to its encoded codon to amino acid sequence and ultimately the protein. Per Zhang, post-translational modifications (such as glycosylation modification), folding, and correct cleavage are essential for proper recombinant protein production for which he suggests the use of cultured Chinese hamster ovary cell as the cell culture media that is to be transfected with the genetically engineered plasmid due to its capacity to allow glycosylation modifications [22]. But efforts should be made to avoid animal models if possible, to reduce costs and bottle neck in recombinant mAb mass production even further. A suggestion may be to improve glycosylation-engineered yeast to obtain the same capacity as Chinese hamster ovary cells. Another suggestion is to try a new larger plant for recombinant mAb production that can allow to be transfected with genetically engineered plasmid for larger recombinant proteins such as the world’s largest single-celled organism, an aquatic algae called Caulerpa taxifolia which can grow up to 6 to 12 inches [46].
• Additional methods of making mAb production more cost effective: In Vitro mAb production in yeast culture: Add a promotor to IVT mRNA in step 5 and after cloning the IVT mRNA in step 6, stimulate transcription of plasmid DNA (pDNA), encode the IVT mRNA, and stimulate translation of IVT mRNA within the yeast culture for mass mAb production within the yeast culture. Provide a medium rich in amino acids necessary for the mAb production. This process is similar to the process of producing foreign protein synthesis in yeast cultures by use of plasmid for encoding DNA as done in past experiments [45,47]. This is also similar to the current recombinant monoclonal antibody production that relies on a known antibody gene from immunized animals or from hybridomas to mass produce in yeast cultures via plasmid genetic engineering and recombinant DNA technology.
• More costly and slower production of monoclonal antibody development is the use of transgenic animal models: Synthesize synthetic DNA, which when transcribed, encodes the united mRNA and inseminate in ovum lamb and proceed to select transgenic progeny to have HOPE monoclonal antibodies secreted in the progeny lamb’s milk [46]. Purification process of the HOPE mAbs with this method may increase cost.
• More costly and slower production of monoclonal antibody development is the use of animal models: Transcribed and synthesized synthetic DNA encodes the united mRNA and genetically engineer in vitro hybridoma and later inseminate in animal models [48]. One can also test if genetically engineered in vitro hybridoma production of HOPE mAb can be done without the use of animal models [49].
In either of these options, it must follow with the extraction of HOPE mAb and quality testing of the pure HOPE mAb. Animal Testing for safety and efficacy of HOPE monoclonal antibodies followed by human trials. One may make a hypothesis that the efficacy of the HOPE monoclonal antibodies should prove to be the same as the selected antibody from step 1 that is if the de novo peptide sequencing algorithms and the series of other computational models predicted the mRNA sequence correctly. If efficacy proves to be less than the selected antibody, review the de novo peptide sequencing algorithms and computational models in decoding the mRNA sequence or the purification mechanisms of the monoclonal antibody. This method was created to make mAb production in yeast culture a more cost-effective mass production method for a number of diseases or tumors that produce antibodies in recovered patients.
HOPE mAbs for infectious diseases: The HOPE method can work with any disease that promotes an antibody response. Although the method only mentions SARS-CoV-2 as its pathogen that can be applied to other viral infections, non-viral pathogens that provoke antibodies in recovered patients might also benefit from the HOPE method. Although for non-viral pathogens neutralizing antibodies do not exist and would require studies to confirm efficacy of attacking the non-viral pathogen with antibodies that are very specific to the pathogen. If specificity of FAB region of antibodies to epitope in non-viral infectious pathogens alone is proven to be as effective as neutralizing antibodies against viral pathogens, HOPE mAb against extracellular infectious pathogens may also be a promising method for therapeutic drug development against antibiotic drug resistant super bugs or pathogens. If shown not to be as effective therapeutically, HOPE mAbs specific can still be used for research and to develop rapid diagnostic tests specific for various non-viral pathogens since the binding capacity in its specificity can still be beneficial even though it might not be an effective treatment option. In this case as a result of not to being introduced to humans, less safety studies will be required for use of HOPE mAbs for diagnostic purposes.
HOPE mAbs and cancer: HOPE mAb for therapeutic purposes in oncological patients for precision and personalized medicine will be harder to prove their safety and efficacy in clinical trials since each HOPE mAb use in a cancer patient will be specific for the neoantigen only found in that patient’s tumor. The studies must be case studies similar to the struggles CAR T cell therapy has had to prove its safety and efficacy over the years.
The HOPE method specific against neoantigens in tumors can create a personalized HOPE mAb for therapeutic and diagnostic purposes in oncological patients that is specific for each patient and is not generalized for the public. While natural antibodies against tumor neoantigens are normally secondary to the body’s primary defense against tumors infiltrating lymphocytes (TILS), the development of precision mAb specific against neoantigens in individual patients is possible with the HOPE method. Mimicking the natural antibodies can be tested to see if it helps prevent metastasis or alter the tumor’s micro environment by increasing the tumor’s immunogenicity as the HOPE mAb bind to the neoantigens. Personalized rapid tests with HOPE mAb for each oncologic patient can be developed to detect any microsatellite tumor cell in biopsies or serum. Once genetic engineering is perfected in the future, similar to engineering B cells, the cancer patient’s individual plasma cell that can be sampled from the patient, may be edited in vitro with the DNA sequence that transcribes the mRNA of HOPE mAb then transfused back into the patient. The genetically engineered plasma cell can then mass produce HOPE mAb within the patient in response to neoantigens of the tumor thus avoiding the need of repeated administration of HOPE mAb specific against the tumor.
Thus the in vitro transcribed RNA (IVT mRNA) vector encoding the united mRNA sequence of the entire genetically engineered monoclonal antibody (mAb), can be delivered to plasma cells in vitro then administered cell therapy in humans similar to how passive immunity mRNA vaccines are being developed to deliver the mRNA sequence of a known antigen [50,51]. The ethical committee would have to decide whether or not to allow emergency use authorization of HOPE mAb in oncologic patients as a drug of last resort if in vitro studies are promising. HOPE mAb for diagnostic use seems more promising for a more rapid commercial use as they may serve to detect tumor micro satellites in the serum or used in biopsy samples and require efficacy studies on HOPE mAbs’ specificity rather than its safety.
HOPE mAbs and bioproteins: HOPE method can also be applied to material science
As the computational models can also be used to reverse engineer bioproteins whenever the bioproteins gene is unknown. With cryo-Em and mass spectrometer data analysis of the bioprotein and the same computational models used above, the bioprotein’s mRNA can be predicted. And steps 5 and 6 in the HOPE method can be done for bioprotein mass production instead of mAb mass production. If the genes of a bioproteins are known the computational model steps in the HOPE method may be skipped. And if the DNA sequence of the bioprotein gene fit into a plasmid mass production can be done via genetic engineering and recombinant DNA technology in yeast cultures. Although as mentioned before another option is the fragment the gene into several plasmids if the gene does not fit within the plasmid or the use of cultures of Chinese hamster ovary cells or Caulerpa taxifolia algae, if the yeast DNA is too small. Ecological safety studies will have to be done prior to releasing the bioproteins produced by the HOPE method into nature to assure its proper decomposition as well as address safety concerns with biodiversity ecosystems.
Discussion
Current mAb production specific against SARS-CoV-2 is a simplified method in comparison to the HOPE method, as current COVID-19 mAb production goes directly to the source of the COVID-19 survivor’s antibodies’ genes in effector B cells or memory B cells, then using genetic engineering and recombinant technology mAb production technology, bypassing the need for computational models as presented in this paper for the HOPE method. But the issue with current mAb production is that most effector B cells are in the bone marrow and not all the memory B cells are in the circulation, thus cannot be collected in serum samples of recovered patients, and any potential potent neutralizing antibodies produced by effector B cells in the bone marrow may be missed opportunities for the development of potent monoclonal antibodies. On the contrary, with the HOPE method these neutralizing antibodies without known genes whose effector B cell cannot be collected can make it possible for diagnostic and treatment mAb development.
To avoid the need for computational models and machine learning steps in the HOPE mAb method, step 1 can be modified by studying more profoundly the available effector B cells in recovered patients instead of the antibodies of recovered patients in step 1. Culturing in vitro the effector B cells and proper identification of each of the samples and their corresponding antibodies to test its efficacy for proper selection of the best neutralizing antibody can be done [52]. Once the best neutralizing antibody is selected the gene of that neutralizing antibody can be obtained from the corresponding effector B cell that produced it to be used in recombinant monoclonal antibody production. A similar process can be done for memory B cells, but memory B cells will first have to be activated via clonal expansion to differentiate into plasma cells that produce antibodies. Followed by the selection process of identifying the best neutralizing antibody that might be used for the develop mAb for diseases and bypass the steps of machine learning, and might be how current COVID-19 mAb are being developed. However, effector B cell samples in the screening might be limited and not capture the corresponding effector B cell of a potent neutralizing antibody. The potential use of the HOPE method in mass production of bioprotein without known corresponding gene and without computational analysis and machine learning is not possible.
Another approach, upon selection of antibody in Step 1, one can go back to human donor to find the effector B cell corresponding to that particular selected antibody. One can avoid the need of computational models and use genetic engineering instead by using CRISPR technology to obtain the corresponding DNA sequence of the antibody in the effector B cell of the selected antibody or the DNA sequence of the FAB region of the membrane bound Ig in a memory B cell’s receptor specific against the pathogen of interest that can be combined with a DNA sequence of a human FC region. The DNA sequences can then be added to a plasmid to incorporate into yeast cultures to cultivate mAb. But identifying the corresponding effector B cell will be difficult. One might have to clone all of the effector B cells to compare each of their antibodies to the selected antibody in Step 1. Thus, will require testing of all the antibodies of the effector B cells in the selected human host to try to match its antibody’s binding capacity to bind to the same epitope that bound to the selected antibody of Step 1. Another point is that the DNA sequence will have the introns if taken from B cells and be a longer DNA sequence that might make it difficult to fit within a plasmid or yeast DNA. Although this is why mammalian cells have been used for recombinant mAbs, perhaps a plant cell culture will be more economical if possible.
Another advantage of completing at least the first step of the HOPE method is to advance the mRNA vaccine production of various diseases. By finding the best neutralizing antibody within a population in step 1 of the HOPE method, one can study the epitope of the antigen it binds to and this can serve as guides to identify potent targets for vaccines. Since this epitope created a strong immune response in the recovered patient, the epitope’s mRNA sequence can be identified then added to mRNA vaccines. Subsequently the patient receiving the mRNA vaccine will produce a similar potent neutralizing antibody as its FAB region would be specific to the same epitope. If proven safe and effective this can be an approach for rapid vaccine development for the next pandemic as well as an approach for vaccine development of various global health diseases.
Conclusion
Worldwide distribution of a safe and effective vaccine to achieve herd immunity can take years to accomplish. The rapid appearance of variants of SARS-CoV-2 has made it difficult to treat with prior monoclonal antibodies, the reverse engineering of antibodies of recovered COVID-19 patients with the HOPE method, might help with finding and attacking epitopes with less history of mutation as well as reduce cost of production. Since HOPE mAbs for therapeutic use will require time due to safety and efficacy purposes, in the meantime, HOPE mAb can be developed for lab research and rapid diagnostic COVID-19 tests in an attempt to reduce mortality due to COVID-19 by increasing diagnostic rapid tests availability. In the future upon completion of the safety and efficacy studies and if COVID-19 still exists endemically, HOPE mAbs specific against SARS-CoV-2 can be used for therapeutic purposes which are essential for immunocompromised individuals in which the vaccine might be ineffective. HOPE mAbs specific against SARS-CoV-2 for therapeutic use will require safety and efficacy studies which might prolong its commercial therapeutic which depending on the evolution of the COVID19 pandemic will determine its worth in developing HOPE mAbs specific for SARSCOV2 for therapeutic use. HOPE mAbs use in research and diagnostic use seems more promising as it can potentially be used more rapidly and will require only binding efficacy tests to mass produce rapid COVID-19 diagnostic tests. HOPE mAbs specific for SARS-CoV-2 development will all depend on if the advancements in computational models can be made on time for the demand if cases of COVID-19 begin to rise once more or if new variants of concern decrease current vaccine or therapeutic efficacy.
Until HOPE mAbs is shown to be safe and effective for therapeutic purposes, the Hope mAbs have the potential of providing rapid in vitro diagnostic tests for various infectious diseases including personalized diagnostic test for oncologic patients. Together with the suggestions made in the Discussion section of this paper, the implementation of step 1 of the HOPE method can be beneficial for the development of mRNA vaccines for various diseases. Thus, the HOPE method has the potential of providing HOPE mAbs for research, diagnostic tests and therapeutic drugs. In part step 1 of the HOPE method can assist in the development of potential mRNA vaccines to diseases that provoke an adaptive immune response thus has high potential value in global health and future pandemic preparation, by identifying the best antibody and identifying the epitope it binds to. The epitope’s mRNA sequence can be obtained and added to mRNA vaccines to induce a strong immune response as seen in recovered patients.
If the HOPE method is taken further beyond the medical field, it may provide a method to mass produce any biological protein (bioproteins) in the field of material science for potential uses in bioengineering, including bioinspired innovations such as mass-producing enzymes for biobatteries and biogels for 3D bioprinting. This may help reduce the use of plastics and make a more sustainable alternative to plastics as well as potential impact in climate change innovations like algae biofuels.
Footnote
The HOPE method was created by Dr. Michellie Hernandez, MD. (Note from the primary author, Dr. Michellie Hernandez: “I shared the idea of the HOPE method publicly during its concept phase in April 2020 to be viewed by researchers globally to start HOPE mAb production globally, as quickly as possible due to the severity of the COVID-19 pandemic.
Later on, I made public the HOPE method more in detail in June 2020 in LinkedIn and the NIH Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) program. I also made it public in worldpulse (Hernandez 2020) and later on in 2021 as I was granted access to ResearchGate with help of a recommendation from a researcher (Hernandez 2021). Even though I had made prior attempts to publish the earlier version of the HOPE method, it was rejected by journals and never published or peer reviewed. The earlier version of the HOPE method is listed in the references and cited as 2021, which was when I was granted access to make it public in ReseachGate and obtained a DOI for proper citation. Recently, I was encouraged to try to publish my research once more for proper peer review and is the reason I have written a more detailed version in this paper along with Deb Bose.”)
Acknowledgement
“I will like to thank Dr. Mary Ruebush, PhD who encouraged me to continue in the early stages of my research.” - Dr. Michellie Hernandez.
Conflicts of Interest
No conflict of interests by any of the authors
Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, et al. (2004) An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nature Medicine 10: 871-875. [ Ref ]
Barderas R, Benito-Peña E (2019) The 2018 Nobel Prize in Chemistry: phage display of peptides and antibodies. Analytical and Bioanalytical Chemistry 411: 2475-2479. [ Ref ]
Payne S (2017) Viruses: from understanding to investigation. Academic Press, Elsevier. [ Ref ]
Sok D, Moldt B, Burton D (2013) SnapShot: Broadly Neutralizing Antibodies. Cell 155: 728-728. [ Ref ]
Nishimura Y, Martin M (2017) Of Mice, Macaques, and Men: Broadly Neutralizing Antibody Immunotherapy for HIV-1. Cell Host & Microbe 22: 207-216. [ Ref ]
Yamin R, Jones A, Hoffmann H, Schäfer A, Kao K, et al. (2021) Fcengineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature 599: 465-470. [ Ref ]
Palm A, Henry C (2019) Remembrance of Things Past: Long-Term B Cell Memory After Infection and Vaccination. Frontiers in Immunology 10. [ Ref ]
Justiz V, Jamal Z, Ramphul K (2023) Immunoglobulin. StatPearls Publishing pending. [ Ref ]
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, et al. (2002) Molecular Biology of the Cell. Garland Science, New York. [ Ref ]
Eisen H (2014) Affinity Enhancement of Antibodies: How Low-Affinity Antibodies Produced Early in Immune Responses Are Followed by High-Affinity Antibodies Later and in Memory B-Cell Responses. Cancer Immunology Research 2: 381-392. [ Ref ]
Gutzeit C, Chen K, Cerutti A (2018) The enigmatic function of IgD: some answers at last. Eur J Immunol 48: 1101-1013. [ Ref ]
Chen K, Cerutti A (2011) The function and regulation of immunoglobulin D. Curr Opin Immunol 23: 345-352. [ Ref ]
Geisberger R, Lamers M, Achatz G (2006) The riddle of the dual expression of IgM and IgD. Immunology 118: 429-437. [ Ref ]
Maity P, Datta M, Nicolò A, Jumaa H (2018) Isotype Specific Assembly of B Cell Antigen Receptors and Synergism With Chemokine Receptor CXCR4. Frontiers in Immunology 9. [ Ref ]
Fera D, Schmidt A, Haynes B, Gao F, Liao H, et al. (2014) Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains. Proceedings of the National Academy of Sciences 111: 10275-10280. [ Ref ]
Parker N, Schneegurt M, Thi Tu A-, Lister P (2022) Microbiology (Chapter: Overview of Specific Adaptive Immunity). OpenStax. 2022. CC BY 4.0. Copyright Rice University, Houston. [ Ref ]
Parker N, Schneegurt M, Tu T, Lister P, et al. (2016) Practical Applications of Monoclonal and Polyclonal Antibodies. Open Stax, Microbiology. 2022 CC 4.0 Copyright Rice University. [ Ref ]
Berzofsky J, Howe S, Olkhanud P (2022) Antigens. Encyclopedia of Infection and Immunity 1: 76-89. [ Ref ]
Sun L, Middleton DR, Wantuch PL, Ozdilek A, Avci FY (2016) Carbohydrates as T-cell antigens with implications in health and disease. Glycobiology 26: 1029-1040. [ Ref ]
Corti D, Purcell L, Snell G, Veesler D (2021) Tackling COVID-19 with neutralizing monoclonal antibodies. Cell 184: 4593-4595. [ Ref ]
Mitra S, Tomar PC (2021) Hybridoma technology; advancements, clinical significance, and future aspects. Journal of Genetic Engineering and Biotechnology 19. [ Ref ]
Zhang J, Shan L, Liang F, Du C, Li J (2022) Strategies and Considerations for Improving Recombinant Antibody Production and Quality in Chinese Hamster Ovary Cells. Frontiers in Bioengineering and Biotechnology 10. [ Ref ]
Frenzel A, Hust M, Schirrmann T (2013) Expression of Recombinant Antibodies. Frontiers in Immunology 4. [ Ref ]
Xia X (2021) Domains and Functions of Spike Protein in SARS-Cov-2 in the Context of Vaccine Design. Viruses 13. [ Ref ]
Hwang Y, Lu R, Su S, Chiang P, Ko S, et al. (2022) Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. Journal of Biomedical Science 29. [ Ref ]
FDA, et al. (2022) FDA Emergency Use Authorization official website. [ Ref ]
US department of Health and Human Services (2022) Public Health Emergency. [ Ref ]
National Institute of Health (2022) NIH COVID19 Treatment Guidelines on SARS-CoV-2 Variants and Susceptibility to Anti-SARS-CoV-2 Monoclonal Antibodies. [ Ref ]
Cooper M, Alder M (2006) The Evolution of Adaptive Immune Systems. Cell 124: 815-822. [ Ref ]
Kirchdoerfer R, Herrin B, Han B, Turnbough C, Cooper M, et al. (2012) Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores. Structure 20: 479-486. [ Ref ]
Hershberg U, Luning Prak E (2015) The analysis of clonal expansions in normal and autoimmune B cell repertoires. Philosophical Transactions of the Royal Society B: Biological Sciences 370. [ Ref ]
Hernandez M (2021) HOPE Monoclonal Antibodies: Genetically engineered monoclonal antibodies via mass spectrometry or cryogenic electron microscopy followed by protein design analysis of antibodies of recovered patients. [ Ref ]
Hernandez M (2020) HOPE Monoclonal Antibodies: Genetically Engineered Monoclonal Antibodies via Mass Spectrometry or Protein Design analysis of antibodies of Recovered patients. By Dr. Michellie Hernandez, MD is licensed under CC BY-SA 4.0. [ Ref ]
Pinto D, Park YJ, Beltramello M (2020) Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583: 290-295. [ Ref ]
Callahan N, Tullman J, Kelman Z, Marino J (2020) Strategies for Development of a Next-Generation Protein Sequencing Platform. Trends in Biochemical Sciences 45: 76-89. [ Ref ]
Tran NH, Zhang X, Xin L, Shan B, Li M (2017) De novo peptide sequencing by deep learning. Proceedings of the National Academy of Sciences 114: 8247-8252. [ Ref ]
Kuhlman B, Bradley P (2019) Advances in protein structure prediction and design. Nature reviews. Molecular cell biology 20: 681-697. [ Ref ]
Lorimer D, Raymond A, Walchli J, Mixon M, Barrow A, et al. (2009) Gene Composer: database software for protein construct design, codon engineering, and gene synthesis. BMC Biotechnology 9. [ Ref ]
Stothard P (2000) The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. BioTechniques 28: 1102-1104. [ Ref ]
Fujimoto MS, Bodily PM, Lyman CA, Jacobsen JA, Clement MJ (2017) Learning the Language of Genes: Representing Global Codon Bias with Deep Language Models. [ Ref ]
Giugno R, Pulvirenti A, Ragusa M, Facciola L, Patelmo L, et al. (2004) Locally sensitive backtranslation based on multiple sequence alignment. IGARSS 2004. 2004 IEEE International Geoscience and Remote Sensing (IEEE Cat. No.04CH37612). [ Ref ]
Ingber D (2022) Human organs-on-chips for disease modelling, drug development and personalized medicine. Nature Reviews Genetics 23: 467-491. [ Ref ]
Goodwin S, McPherson J, McCombie WR (2016) Coming of age: ten years of next-generation sequencing technologies. Nature Reviews Genetics 17: 333-351. [ Ref ]
Schlake T, Thess A, Thran M, Jordan I (2018) mRNA as novel technology for passive munotherapy. Cellular and Molecular Life Sciences 76: 301- 328. [ Ref ]
Ridder R, Schmitz R, Legay F, Gram H (1995) Generation of Rabbit Monoclonal Antibody Fragments from a Combinatorial Phage Display Library and Their Production in the Yeast Pichia pastoris. Nature Biotechnology 13: 255-260. [ Ref ]
Donald Danforth Plant Science Center (2015) Structure of world’s largest single cell is reflected at the molecular level. ScienceDaily. [ Ref ]
Griffiths AJF (2000) An Introduction to Genetic Analysis; Chapter: Recombinant DNA technology in eukaryotes. WH Freeman, New York. [ Ref ]
Potter M, Chang P, et al. (2007) Artificial Cells, Cell Engineering and Therapy. Woodhead Publishing Series in Biomaterials. [ Ref ]
Van Hoecke L, Roose K (2019) How mRNA therapeutics are entering the monoclonal antibody field. Journal of Translational Medicine 17. [ Ref ]
Pardi N, Hogan M, Porter F, Weissman D (2018) mRNA vaccines — a new era in vaccinology. Nature Reviews Drug Discovery 17: 261-279. [ Ref ]
Pardi N, Secreto A, Shan X, Debonera F, Glover J, et al. (2017) Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nature Communications 8. [ Ref ]
Andreano E, Nicastri E, Paciello I, Pileri P, Manganaro N, et al. (2021) Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell 184: 1821-1835. [ Ref ]