Journal Name: Journal of Biomedical Research and Reviews
Article Type: Research
Received date: 13 February, 2019
Accepted date: 18 February, 2019
Published date: 2021-03-21
Citation: Dubytska LP (2019) Borrelia burgdorferi 297 bmpA Encode the mRNA that Contains ORF for a Leader Peptide that Regulates bmpA Gene Expression. J Biomed Res Rev Vol: 2, Issu: 1 (34-44).
Copyright: © 2019 Dubytska LP. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
The Bmp proteins are highly conserved proteins with no well established functions in B. burgdorferi sensu lato and are immunogenic. It was reported that four genes from this cluster bmpD-bmpC-bmpA-bmpB are expressed in vitro as monocistronic and polycistronic messages.
Evidence is presented in this report that bmpA mRNA contains two ribosome binding sites (SD) separated by 90 bases pairs. The SD1 precedes a small 32 amino acid ORF - leader peptide (BmpAL). The SD2 is the RBS for 342 amino acids BmpA. The bmpAL and bmpA ORFs in B.burgdorferi 297 overlap by eight base pairs suggesting that two proteins can be co-regulated. First five codons in the leader peptide and “-GGG-“ in SD2 are rarely used in Borrelia, suggesting that they can regulate BmpAL and BmpA expression. Deletion of SD1 in the leader sequence, or introducing a stop codon immediately before SD2 leads to increased BmpA::GFP expression in B.burgdorferi 297 that contains bmpA::gfp translational fusion on the plasmid. In B. garinii G25 and B. afzelii IP3 the leader sequence is in frame with bmpA, and as result, in B. afzelii IP3 BmpA is expressed as the higher molecular weight protein compared to BmpAs of B. burgdorferi 297 and B. afzelii DK7.
Keywords
Leader; bmpA; Translation regulation.
Abstract
The Bmp proteins are highly conserved proteins with no well established functions in B. burgdorferi sensu lato and are immunogenic. It was reported that four genes from this cluster bmpD-bmpC-bmpA-bmpB are expressed in vitro as monocistronic and polycistronic messages.
Evidence is presented in this report that bmpA mRNA contains two ribosome binding sites (SD) separated by 90 bases pairs. The SD1 precedes a small 32 amino acid ORF - leader peptide (BmpAL). The SD2 is the RBS for 342 amino acids BmpA. The bmpAL and bmpA ORFs in B.burgdorferi 297 overlap by eight base pairs suggesting that two proteins can be co-regulated. First five codons in the leader peptide and “-GGG-“ in SD2 are rarely used in Borrelia, suggesting that they can regulate BmpAL and BmpA expression. Deletion of SD1 in the leader sequence, or introducing a stop codon immediately before SD2 leads to increased BmpA::GFP expression in B.burgdorferi 297 that contains bmpA::gfp translational fusion on the plasmid. In B. garinii G25 and B. afzelii IP3 the leader sequence is in frame with bmpA, and as result, in B. afzelii IP3 BmpA is expressed as the higher molecular weight protein compared to BmpAs of B. burgdorferi 297 and B. afzelii DK7.
Keywords
Leader; bmpA; Translation regulation.
Introduction
Borrelia burgdorferi, the spirochetal bacterium that causes the tickborne infection called Lyme disease [1,2].B. burgdorferigenome contains approximately 1000 chromosomal and 400 plasmid genes [3] but only a few homologs to regulatory genes, sigma factors and one rho terminator factor [3]. In addition, Borrelia has genes and gene families that do not share homology with genes of other bacteria [3] suggesting that B. burgdorferi may have different mechanisms to control gene expression.
Evolutionary selected systems of virulence gene regulation allow coordinated gene expression that is based on the temporal and special requirements of host niches. Global regulation of virulence genes is a common strategy of bacterial pathogens to overcome the complexity of innate host defenses [4-11]. In addition to a global regulatory system, prokaryotes can employ non-global mechanisms of virulence gene regulation. They include expression of non-coding RNAs [12,13], effects on mRNA secondary structure that forms terminator/anti-terminator structure [14-16] and affects mRNA stability [17] as well as the differential efficiency of ribosomal binding [18,19].
The bmp gene cluster ofB. burgdorferiis located in the chromosome and encodes lipoproteins with high amino acid homology, that are expressed in vivo and are immunogenic [20-22]. In humans and animals antibodies against one of the members of this family, BmpA (formerly p39), appear early during infection [21].B. burgdorferiwith bmpA or bmpB deletions is unable to persist in mouse joint tissues [23]. The BmpA can also stimulate the production of inflammatory cytokines in human and murine lymphocytes, indicating an important role of BmpA in the maintenance of mammalian infection [23].
According to Dobricova et al. [24], four bmp genes are expressed in vitro and constitute two transcriptional units with a complex pattern of transcription, including alternative monocistronic and polycistronic messages. One unit contains bmpD, and the second unit includes bmpC, bmpA and bmpB. Moreover, promoters were identified for bmpD, bmpC and bmpA, but not for bmpB. The bmpC is always expressed as a polycistronic message with bmpA, and bmpA can transcribe as individual mRNA and as bicistronic bmpAbmpB. According to Ramamoorthy et al. [25] expression from the bmpA-bmpB operon results in three distinct transcripts bmpA, bmpA-bmpB and bmpA truncated bmpB. In addition, the conservation of bpm genes within the B. burgdurferi sensu lato complex and the presence of orthologs in Treponema pallidium and numerous other bacteria suggest that these proteins can play an essential physiological role.
Unusual genetical structure Bmp genes and pattern of their expression may indicate specific regulatory mechanisms that are involved in the expression of these genes. To uncover some of the questions about BmpA expression and regulation, we investigate bmpA transcript and role of the leader sequence (bmpAL) on BmpA expression.
Materials and Methods
Bacterial strains and medium
E. coli DH5α (New England BioLabs, Beverly, MA) and E. coli TOP10 were grown in Luria-Bertani (LB) broth or plates (Gibco-BRL, Gaithersburg, MD). TheB. burgdorferi279 [26] was grown in BSK-H medium (Sigma, St. Louis, MO.) with 6% rabbit serum (Sigma, St. Louis, MO). Appropriate antibiotics were added when specified.
DNA manipulations were performed by standard methods [27]. Restriction enzymes were obtained from New England BioLabs, Beverly, MA. Total DNA was purified from bacterial cultures using High Pure PCR Template Preparation kit (Roche, Mannheim, Germany), DNA fragment and PCR product purification was done using QIAquick Gel Extraction kit (Qiagen, Valencia, California.); all methods were performed according to the manufacturers’ instructions. Constructions were done as previously described [28] by using long PCR. Oligonucleotide primers used in this work were purchased from Integrated DNA Technologies, Skokie, Illinois. All constructs were confirmed by PCR amplification with appropriate primers (Table 1) and DNA sequence analysis of amplicons.
Table 1: Primers used in this work.
| Name | Sequence 5/-3/ |
|---|---|
| P3c | Tagctttgtttgtaaaatagtttatgagtaaaggagaagaacttttcac |
| P2c | Gtgaaagttcttctcctttactcataaactattttacaaacaaagcta |
| P1a,b,c,d,e,g,h,I,j,f | Attacacggggtaccccggcacctcaaaatgttattacttcaata |
| P2a | tgggacaactccagtgaaaagttcttctcctttcatcataaactatttcccctttacaaacaaagctatatt |
| P4a,b,c,d,e,g,h,I,j,f | tcagcatgcttatttgtatagttcatccatgccatgtgtaatcccagc |
| P3a | aatatagctttgtttgtaaaggggaaatagtttatgatgaaaggagaagaacttttcactggagttgtccca |
| P3b,e | gaaaataaaataataaaaattattgttcctgatagtgaatatgc |
| P2b,e | caataatttttattattttattttctagatcaataacttcatcaaccaac |
| P3f | gaaaataaaataataagtggagaaattattgagtaaaggagaagaac |
| P2f | gttcttctcctttactcaataatttctccacttattattttattttc |
| P3d | gttgttgattttgctgtagcgtaaaggagaagaacttttc |
| P2d | gaaaagttcttctcctttacgctcaagcaaaatcaacaac |
| P3h | gcatttgatttatttaaatcaaagttattaactacttaaatatagc |
| P2h | gctatatttaagtagttaataactttgatttaaataaatc |
| P3g | gtttgtaaaggggaaatagtttatgaataaaggagaagaacttttc |
| P2g | gaaaagttcttctcctttattcataaactatttcccctttacaaac |
| P3i | ttgttcctgaatagtgaatatgcatttgatttatttaaatcaaagttaaaactacttaaatatagc |
| P2i | gctatatttaagtagttttaactttgatttaaataaatcaaatgcatattcactattcaggaac |
| P3J | caaagttataaactacttaatatagctttgtttgtaaaggggaaatag |
| P2J | ctatttcccctttacaaacaaagctatattaagtagtttataactttg |
Constructs: a) bmpAL::gfp |
|
BmpAL-Gfp fusions and BmpL mutations construction
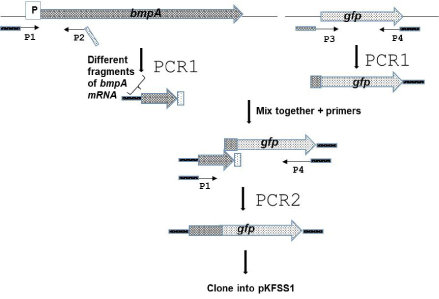
The strategy for constructing the Gfp fusions is shown in Figure 1. Different lengths of bmpA mRNA sequence was amplified fromB. burgdorferi297 total DNA with a gene-specific forward primer, P1, that annealed at least 190 bp upstream from the translational start codon in order to incorporate the native promoter and included a linker containing a specific restriction enzyme (RE) site to facilitate cloning. Primer P1 was paired with the reverse primer, P2, which included a linker that contained 25 to 30 bp gfp. The Gfp amplified from pCE320 [29] with primer P3, which included 25 to 30 bp of the specific BmpA sequence and primer P4, which included an in-frame stop codon and another RE site.
Figure 1: Schematic description of the construction process of theB. burgdorferi297 BmpAL-FGP constructs. The different BmpAL-GFP constructs were made by truncating bmpAL and bmpA, as well as introducing deletions of SD1, SD2 and leader mutations in the primers.
Deletions of SD1 or SD2, stop codons and leader sequence mutations were introduced in the primers and incorporated in the constructs by PCR. Constructs that contain both SDs and has no mutations were created first and then were used as a template to generate constructs menschen above.
Primers used to amplify the GFP and the individual BmpA sequences are listed in table 1. To produce the fusion constructs, each BmpA fragment or mutant and GFP amplicons were mixed and amplified using P1 and P4 primers.
The PCR amplification parameters for all constructs in this work were as follows: denaturation for 2 min at 94°C for one cycle, followed by 38 cycles of 94°C for 10 s, 53°C for 10 s, 72°C for 2 min, and a final extension at 68°C for 5 min. The resulting PCR product was purified and cloned into pCR2.1-TOPO and subsequently electroporated into E.coli TOP10. Plasmid DNA from electroporants selected on Luria-Bertani agar plates with kanamycin or ampicillin (according to manufacture instruction) was purified. Then each construct was excised and subcloned into pKFSS1 [30]. DNA fragments containing cloned constructs in all structures were confirmed by DNA sequencing.
All constructions are located under native B. burgdorferi 297 BmpA promoter and contain different length of BmpA mRNAsequnse. Construct bmpAL::gfp contains mRNA bmpA sequence from -190 base pair (bp) to bmpA starting codon (-AUG-) and gfp under this start codon. Constructs bmpAL(ΔSD1)::gfp and bmpAL(ΔSD1)::gfp differ from first one by deletion of SD1 (-GTGGAG-) and SD2 (-AGGGGA-), respectively. In constructs bmpAL(33bpbmpA)::gfp, gfp starts after 33 bp of bmpA gene respectively, and in bmpAL SD1::gfp contains gfp starts after bmpAL start codon -UUG-. In the bmpAL stop::gfp, gfp is fused after leader peptide stop codon.
TheB. burgdorferielectroporation.B. burgdorferi297 at mid-log phase (1-2 x 107 cells/ml) was electroporated with 10 to 30 μg of recombinant plasmid DNA. After overnight recovery, cells were diluted to 107 cells/ml and distributed into 96 micro-well plates (Corning Incorporated, Corning, N.Y.) containing BSK-H media with 70-100 μg/ml of streptomycin for selection of clones containing recombinant plasmid. After 10-15 days DNA ofB. burgdorfericells growing in these microwells was checked for the presence of the plasmid by fluorescence and by PCR. The DNA of streptomycin resistant colonies was extracted using High Pure PCR Template Preparation Kit (Roche Diagnostics Corporation, Indianapolis, IN) and analyzed by PCR for the presence of the appropriate construct with specific primers (Table 1).
Detection of GFP and BmpA by immunoblotting
E. coli DH5α andB. burgdorferi297 total proteins were extracted from 1-2x107 cells/ml by lysing them in Laemmle buffer. Protein lysates were analyzed by SDS-PAGE followed by silver stain or immunoblotting using rabbit anti- GFP (Invitrogen, Eugene, Oregon, USA) or anti-BmpA polyclonal antibody. Immunoblots were developed using ECF Western Blotting Kit according to the manufacturer’s instructions (Amersham Biosciences, Piscataway, N.J.), and detected using a Storm 860 PhosphorImager and ImageQuaNT software (Molecular Dynamics, Sunnyvale, CA).
Flow cytometry analysis
Aliquots from three independent experiments, containing E. coli at OD06=08 and 1x108B. burgdorferiB31 and its derivatives containing GFP in pKFSS1 or TOPO were washed with PBS and analyzed on a FACS scan flow cytometer (Becton Dickinson, Mountain Lake, Calif.) using CELLQUEST 3.2 (Becton Dickinson).
Microscopic analysis
Cultures of E. coli andB. burgdorferi279 that contain different constructs (106 cells/ml) were examined by fluorescence microscopy to detect the fluorescence.
Results
Analyze the bmpA gene
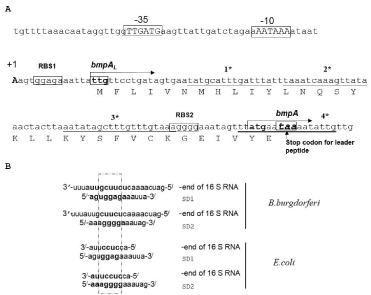
The transcription start site of the bmpA gene is set at -105bp position relative to its translational initiation codon and is located within the coding sequence of the bmpC gene (60 bases upstream from the bmpC stop codon).
There are several palindromic sequences in the bmpAL present, suggesting that bmpAL can also form complicated secondary structures and two purine reach regions that can serve as a ribosome binding site (RBS) [31-33]. Moreover, the ORFs of a leader peptide and BmpA can form two different frames and contain a stop codon for a BmpAL that can overlap with the start codon of BmpA (Figure 2A) suggesting that these two proteins can be co-expressed and co-regulated. Introducing mutations to the palindromes, in the way that they change the mRNA secondary structure but not affect the amino acid sequence, does not affect BmpAL and BmpA expression (data are not shown).
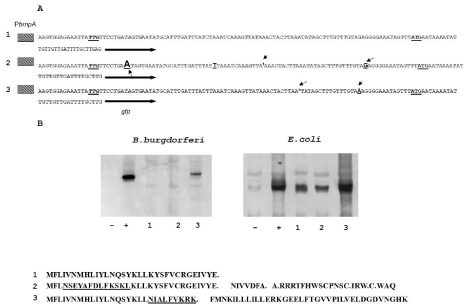
Figure 2: The nucleotide sequence of the 5’-flanking regions of the bmpA gene. A. Transcription initiation site is indicated as +1. Putative bmpA leader peptide (bmpAL) is also shown. Translational initiation codons for bmpAL and bmpA are boxed; stop codon for leader peptide is indicated by the arrow. The invert rapids are indicated by the star. B. Alignment of the B.burgdorferi SD sequences with 3/ end of 16S rRNA. The two Shine – Dalgarno (SD) sequences found in the bmpA mRNA can base pair with the 3/ end of the 16S rRNA.
The SD1 sequence is -GGAG- with a spacing between this SD1 and the initiation codon -UUG- of 10 bp as counted from the first G in SD1 (Figure 2A). The spacing between SD2 (-AGGGGA-) and the initiation codon -AUG- is 12 bp counted from the second G at position 3 in SD2 (Figure 2A). Both SDs in bmpA mRNA can pair with the 3’ end of the 16S rRNA (Figure 2B) [31] suggesting that both SDs can be functional in B. burgdorferi .
Determination of RBS for bmpA
Careful sequence analysis of the bmpA gene shows the presence of two potential SDs. The first SD1 (-GGAG-) starts at nucleotide position +4 counting from transcription start codon and is close to an alternative start codon -UUG-. The sequence (-GGAG-) is classical SD sequence for many species of bacteria and was found approximately in 43% genes of B. burgdorferi when searched in PubMed database.
The distance between (-GGAG-) and the translation start codon for bmpA is 100 bp. The effects of SD spacing, distance between SD and the initiation codon, variation in SD sequences and the effects of other alternative translational start sites are well studied. The excessively long, or short spacing between the SD and the initiation codon may abolish or limit efficient translation initiation [34,35].
The second SD (-AGGGGA-) is located at nucleotide position 90 counting from +1 and 14 nucleotides from the described translation start codon -AUG- for bmpA. Sequence –AGGGGA- is less common as an SD and does not appear as an SD sequence in the database for B. burgdorferi . Moreover, two of the predicted SDs can pare with 16S rRNA (Figure 2B).
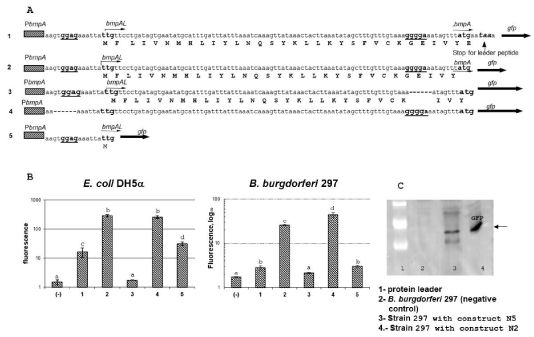
To verify -GGAG- or -AGGGGA- is an SD for BmpA we made several constructs that differ only in bmpAL. One, of these, contain the bmpA promotor bmpAL and the translation start codon -AUG- of bmpA fused to gfp (bmpAL::gfp). A second differs from the first one only by a deletion in the SD2 (-AGGGGA-) sequence ((bmpAL(ΔSD2)::gfp)), and the third construct contains deletion of the SD1 (-GGAG-) sequence (bmpAL(ΔSD1)::gfp). Expression of gfp was studied in both E. coli which served as a model microorganism as well as in B.burgdorferi strain 297.
The results of flow cytometric analysis are presented in figure 3. In the plasmid that harbored bmpAL(ΔSD2)::gfp (Figure 3. line 2) fluorescence in E. coli and B. burgdorferi strains were not detected. In opposite, deletion of SD1 did not abolish the GFP expression (Figure 3. line 3), and in E. coli GFP expression was at the same level as in construct bmpAL::gfp that contains both SD sites. At the same time, in B.burgdirferi 297 GFP expression was approximately twice higher comper to GFP expression from construct bmpAL::gfp. This data suggests that -AGGGGA- is indeed an SD site of BmpA. Moreover, the facts that both SDs (-GGAGand -AGGGGA-) can pair with 3’ end of 16S rRNA of B. burgdorferi and may form the translation initiation region (SD, initiator codon, and a spacer region) suggest that both SDs can be active (Figure 2A, 2B).
Figure 3: Expression of GFP in recombinant strains. A. Sequences of different constructs fused to gfp. 1. The bmpALstop::gfp construct, that contains bmpA promoter bmpA leader from +1 to the BmpA stop codon and gfp fused in frame with BmpA start codon. 2. The bmpAL::gfp construct, differ from construct one by fusion gfp directly to the bmpA start codon -ATG-. 3. The bmpAL(ΔSD2)::gfp construct, differ from the second construct by deletion of SD2 (-AGGGGA-). 4. The bmpAL(ΔSD1)::gfp construct, differ from the second construct by deletion of SD1 (-TGGAGA-). 5. The bmpAL SD1::gfp, contains PbmpA and bmpAL from +1 to -UUG- (start codon for leader peptide) fused in frame to gfp. B. Expression of GFP detected by flow cytometry in E. coli and B.burgdorferi recombinant strains. Level of GFP expression from the constructs: 1) bmpALstop::gfp; 2) bmpAL::gfp; 3) bmpAL(ΔSD2)::gfp; 4) bmpAL(ΔSD1)::gfp; 5) bmpAL SD1::gfp. Statistical analysis was conducted using 1-way ANOVA followed by Tukey’s post hoc test for pairwise comparisons. Data are mean ± SD of 3 replicates; columns with the same letters are not significantly different (p < 0.05). C. Representative Immunoblot for detection of GFP expression in recombinant strains E. coli and B.burgdorferi.
Detection of the leader peptide
To verify that SD1 (-GGAG-) is active and can form translation initiation region (TIR) together with -UUG-, we constructed plasmid in which gfp was fused with the first start codon -UUG- after predicted SD1- (-GGAG-) (Fig.3. line 4). This plasmid allowed GFP production from the start codon for leader peptide under the control of native bmpA promoter (PbmpA) only if -GGAG- plays the role as an SD sate and -UUG- as a start codon. Expression of GFP from this construct was studied in E. coli andB. burgdorferiand compared with expression of GFP from the plasmid that contains both SDs (Figure 3. line 1).
Flow cytometry analysis showed expression of GFP in E. coli andB. burgdorferifrom the plasmid that harbored gfp fused in frame to a start codon of the leader peptide. Expression of GFP from this construct was also detected by western blotting (data not shown). This result indicates that the BmpAL is translated from -UUG- start codon using -GGAG- as SD1. The low-level expression may be explained by rearly used start codon -UUG-.
Expression of leader peptide inhibits bmpA gene expression
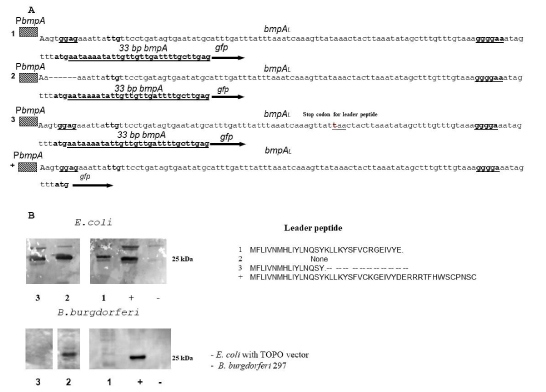
To detect that leader peptide expression has any effect on BmpA expression we created a construct that contains PbmpA, bmpAL, and 33 bp of bmpA fused in frame with gfp protein. The second construct was created from a first one by deletion of sequence -GGAG- that corresponds to SD1 (bmpAL(ΔSD1)33bpbmpA::gfp). Expression of GFP was significantly higher in the E. coli andB. burgdorferistrains that contain SD1 deletion (bmpAL(ΔSD1)33bpbmpAgfp) compared to strains that contain both SDs bmpAL 33bpbmpAgfp construct (Figure. 4A, 4B).
Figure 4: Leader peptide expression inhibits expression of BmpA. A. Sequence of different constructs fused in frame with gfp. 1. bmpAL33bpbmpA::gfp construct, that contain bmpA promoter, bmpAL, 33bp of bmpA fused in frame with gfp. 2. The bmpAL(ΔSD1)33bpbmpA::gfp construct, differs from construct one by deletion of SD1 (-TGGAGA-). 3. The bmpALochre1733bpbmpA::gfp construct, that contains bmpA promoter bmpA leader with the stop codon in position 17, 33bp of bmpA fused in frame with gfp.4. The bmpAL::gfp construct, contains bmpA promoter bmpAL leader from +1 to the BmpA start codon that fused in frame with gfp. The bmpAL::gfp was used as positive control. B. Western blot analyses expression GFP from different construct is described above.
B. burgdorferi bmpA monocistronic message contains two SDs. The fact that SD2 is active even when SD1 is deleted suggests that SD2 is not translationally coupled to SD1 by secondary structure, moreover elevated level of expression in the case where SD1 was removed compare to the construct that contains both SDs suggest that translation of BmpAL inhibits BmpA translation (Figure 3 and Figure 4). This effect was not detected in E. coli strains and can be explained by stronger pairing of 16sRNA with SD2 compare to SD1 (Figure 3B).
To verify that stop codon for leader peptide plays any role in regulation of the upstream located gene, we created a construct that contains entire bmpAL including stop codon and GFP fused in frame with -AUG- of the bmpA gene (bmpAL stop::gfp). The expression of GFP was detected by flow cytometry (Figure 3). Presence of stop codon significantly inhibited gfp translation, compare to construct were gfp was fused directly to a start codon of bmpA. Moreover, as we expected, according to ribosome pairing with SD in E. coli and B.burgdorferi, the effect was more noticeable in B. burgdorferi compare to E.coli, suggesting that stop codon of BmpAL plays significant role in the expression of bmpA gene.
We also introduced stop codon inside of the leader peptide (Figure 4. construct 3). Western blot analysis shows expression of GFP in this construct only in E. coli, and not in B.burgdorferi (Figure 4B).
Thus, our results demonstrate that SD2 is not translationally coupled to SD1 by secondary structure, translation from SD2 does not require SD1, and translation from SD1 inhibits translation from SD2.
Comparison of bmpAL
B.burgdorferi ORF for bmpAL encodes 32 amino acids leader peptide with molecular weight 3882.67 Daltons. It contains three strongly basic (+) amino acids (K,R), two strongly acidic (-) amino acids (D,E), fourteen hydrophobic amino acids (A,I,L,F,W,V), and ten polar amino acids (N,C,Q,S,T,Y). The Isoelectric Point of this peptide is 8.178, 1.044 Charge at Ph 7.0. Nucleotide sequence for bmpAL contains % A+T = 76.77% C+G = 23.23% where % A = 36.36; % G = 16.16; % T = 40.40; % C = 7.07.
The BmpAL amino acid sequence shows strong similarity to other species of Borrelia leader peptide but we do not find homology to another bacterial leader peptides. First 19 amino acids are strong conservative (Figure 5). Inside of this conservative region located 5 leu codons and 3 of them rarely used in Borrelia, suggesting that they can play a regulatory role.
Figure 5: Sequence alignment of the leader peptides from different Borrelia species. Sequences alignments were done using DNA star. Conservative amino acids are boxed in the DNA sequence and red in amino acids sequence. Stop codon is indicated by a star and rarely used codons-bold underline.
Moreover, BmpAL amino acid sequence also has significant differences between Borrelia species.B. burgdorferistrains 297, N40, B31, BL206 have conservative 32 amino acids leader peptide with stop codon located two nucleotides after start codon for bmpA. The B. bissettii 25015 and B. andersonii 21038 also have 32 bp leader peptide but different in amino acids in the variable region.
B. burgdorferi SH-2-82 andB. burgdorferiBTO1 have shorter-28-amino acids peptide. B. afzelii PKO and B. Garini Pbi have 24 leader peptide, and B. afzelii IP3 or B. garini G25 has leader peptide in frame with bmpA. This data may suggest strain-depenmdent differences in bmpA regulation and expression. For example, B. afzelii IP3 or B. garini G25 can use the SD1 or SD2 for expression of BmpA. It can contain two BmpA products with and without the leader sequence. At the same tame expression bmpA in strains B. burgdorferi SH-2-82,B. burgdorferiBTO1, B. afzelii PKO and B. Garini Pbi can be reinitiated from ORF started from SD1. It is not clear if these phenomena have a biological importance.
The variable sequence of the leader peptide can be important for the inhibition of bmpA translation
Two frameshifting mutations were introduced to the bmpAL to examine role of conservative and variable parts of the bmpAL on bmpA translation. The first two-point mutations alter amino acid sequence between bmpAL 4aa and 16 aa. Expression of GFP in resulting mutant does not differ from wild-type bmpAL. At the same tame a change in amino acids sequence of variable part (deletion -A- in codon 20) significantly increases GFP expression (Figure 6). Moreover, in this case, BmpAL is shorter similarly to B. burgdorferi Sh-2-82 where stop codon located immediately before SD2, indicating that translation at SD2 can reinitiate from ORF started at SD1
Figure 6: The GFP expression from recombinant strains containing constructs with mutations in conservative and variable part of bmpAL. A. Sequence of different constructs fused to gfp that was used for this study: 1. bmpAL33bpbmpAgfp construct that contains bmpA promoter, bmpAL, 33bp of bmpA and gfp fused in frame to bmpA. 2. The bmpALmutatedconserve33bpbmpA::gfp construct difer from the first one by insertion of -A- in position 25 and deletion of -Tin position 62. 3. The bmpALmutatedvariable33bpbmpA::gfp construct, contains deletion of -A- in position 74. B. Expression of GFP detected by western blotting in E.coli and B. burgdorferi . Recombinant strains that contain: 1. bmpAl33bpbmpAgfp; 2. bmpALmutatedconserve::gfp; 3. bmpALmutated variable::gfp.
Working model
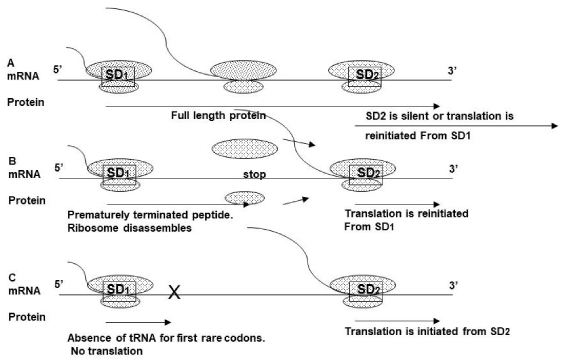
Based on results, described above we proposed a model similar to the models described for E. coli [36,37] and eukaryotic protein-encoding genes that contain upstream ORFs [38] (Figure 7). In strain 297, ribosome proceeds starting from an SD1, it then overrides the SD2, so SD2 site becomes silent (A). In B. afzeliiPKo, B. garini PBi, B. burgdorferi BTO1 the ribosome can reinitiate translation from ORF starting from SD1 (B). In B. garini G25, B. afzelii IP3 SD2 leader peptide fused in frame with the bmpA gene. In this case, two products are possible. First one contains BmpA protein together with leader peptide, another one only BmpA. To test this hypothesis, we performd the western blotting onB. burgdorferi297, N40 and B. afzelii IP3 (Figure 8). Data indicates that BmpA of B. afzelii is slightly larger compared with BmpA ofB. burgdorferi297 and N40, confirming our hypotheses.
Figure 7: Working model. A. In 297 wild-type ribosomes that began translation from SD1 pausing at -GGG- and at stop codon, preventing second ribosome polymerization at SD2. Ribosomes also can re-initiate translation from SD1. B. In strains with a stop codon before SD2 the translation from SD1 is terminated, and ribosome can reinitiate translation at SD2. C. Translation initiation from SD2 when SD1 is abolished.
Figure 8: Expression of BmpA from differentB. burgdorferistrains. A. The BmpAL amino acid sequences ofB. burgdorferi297 (2), B. garini G25 (3), B. afzelii IP3 (4). Amino acids coded by rarely used codons, bold underline. B. Immunoblotting of BmpA expression from different B. burgdorferei strains.
The lower level of translation from constructs that contain both SDs compare wit the same constructs that contain only SD2 can be explained by bmpAL sequence. The first four codons of leader peptide are rare for B. burgdorferi , suggesting that ribosome may translate this region slower. Another fact, that leader peptide inside of conservative region contains five leucine (Leu), codons and three of them are rarely used for B. burgdorferi , suggest that they can be involved in regulation of leader peptide expression. Moreover, another rare codon -GGG- in B. burgdorferi bmpAL is located in SD2 region and can slow down ribosome movement, covering SD2 and inhibiting polymerization second ribosome and translation from SD2. We also do not exclude that additional RNA binding factors or secondary structure of 5’RNA may also play a role in the regulation of leader peptide expression.
Discussion and Conclusion
Post-transcription regulation of gene expression is a key mechanism by which cells and organisms can rapidly change their gene expression in response to internal or external stimuli. Expression of all genes is regulated at multiple post-transcriptional steps including mRNA stability, and translation of mRNA. Translational regulation at the initiation step can be mediated via different cis-acting elements present in the 5′RNA leader sequence, such as the secondary structure of the 5’ RNA and upstream open reading frames (uORFs). The uORFs can significantly change protein expression levels by interfering with the efficiency of translation initiation of the downstream ORF [38,39], indicating that they can control protein synthesis.
Taken together, this data indicates presence of two translation initiation regions in bmpA mRNA. The SD2 is active only when SD1 is silent. 16S rRNA in the 30S ribosomal subunits plays a significant role in selecting the translational start site [32,33]. In most mRNA 4 or 5 bp SD interaction is strong enough to mediate efficient translation [40]. A stronger than regular SD interaction does help, however, when the start codon is not -AUG-, or when the initiation site is masked by secondary structure [41]. A/U –rich initiation site that forms unstable secondary structure might require no SD interaction at all [42].
Stronger base pairing of SD1 sequence with 16S rRNA (8 bp) compare with SD2 (5 bp) inB. burgdorferi(Figure 2B), suggests that ribosome should polymerize more efficiently in SD1. In E. coli the pairing 16S rRNA to SD1 and SD2 is opposite 4 and 6 bp, correspondingly. This fact indicates that in E. coli the ribosome is polymerized more efficiently in SD2. The -UUG- uses about 3% of the start codons in E. coli and it is also rare start codon for a B. burgdorferi . The -AUG- start codon is preferred via pairing with the anticodon (-UAC-) in fMet-tRNA. Weaker pairing is part of the reason for less efficient translation when -GUG- or -UUG- is used as a start codon. In E. coli translation from these codons are 8 times less efficient than from -AUG- [43]. Therefore, even when SD1 have more bases pairing with 16S rRNA in B. burgdorferi the translation of the BmpAL is reduced by using -UUG- as the start codon.
Many bacterial genes are parts of polycistronic operons [44-47]. Translation coupling and re-initiation are important for the expression of functionally related proteins from polycistronic operons [48,49]. The cistrons of some translationally coupled messages do not have an independent SD, and in this case, the stop codon of upstream cistron and the initiator codon of the downstream cistron overlap. A functional ribosome in stop codon for the first peptide reinitiates translation of downstream cistron instead of getting disassembled. A defect in translation from the first cistron abolishes the translation from the downstream cistron [50].B. burgdorferibmpA is transcribed as a monocistronic mRNA that contains two SDs, and as polycistrons with bmpCbmpA and bmpAbmpB. Removing the SD1 from this mRNA increases translation of GFP or BmpA in E. coli andB. burgdorferisuggesting that SD2 is not translationally coupled to SD1 by secondary structure (Figure 3,4,6).
The size of the ribosome is 25nm, indicating that one ribosome may occupy the space approximately 10- 20 aa [51]. When ribosome begins translation from SD1, another ribosome has enough space for polymerization and translation from SD2. But in the absence of translation termination, the ribosomes from SD1 move forward and cover the area necessary for ribosome polymerization at SD2.
The rate at which elongating ribosome translates through ORF is codon-specific and in E. coli differ from 5-21 codons per second. The ribosome can stall during translation elongation in rare or termination codons, creating a blockade to addition ribosome. The ribosome stalling is also involved in positive regulation of translation [51-53].
The first four amino acids in the leader are rare codons and may regulate translation efficiently from SD1. When ribosome occupies two leader codons for Leu, the ribosome progressively encroaches on the space needed for a second ribosome to initiate at SD2. The fact, that bmpAL contains codons that are used less efficiently in Borrelia may also be the reason for a slower translation of bmpAL. Finally, the rare codons for Cys (-UGU-) in the variable part of bmpAL and Gly (-GGG-) inside of SD2 as well as the bmpAL stop codon are the reasons of slower ribosome translation from SD1 that interfere with the second ribosome polymerization at SD2 [53].
When native bmpAL together with a stop codon was present in the derivative plasmids, GFP expression was significantly lower in E. coli and not detected or detected on the low level inB. burgdorfericompare with similar constructs that do not have the stop codon (Figure 3). GFP translation from the construct (Figure 6) in which the stop codon is located before the SD2, was significantly higher compared with translation from construct containing native bmpAL (Figure 7). Moreover, the aa sequence of this construct reminds the sequence of aB. burgdorferiSh-2-82 andB. burgdorferiBTO1. This data suggests that translation from SD2 can happen de novo or by re-initiation from SD1.
Lyme disease patients, having Borrelia burgdorferi infection, show variety of clinical evidences from asymptomatic infection to chronic arthritis. The most common clinical sign of infection is an erythema migrans, caused by a cutaneous B. burgdorferi infection [54,55]. Approximately 5% of untreated patients will develop carditis (e.g., heart block), about 10% will develop neurologic manifestations such as meningitis, cranial nerve palsy or radiculopathy, about 60% will develop arthritis [54], and about 20% of patients do not produce any subsequent clinical manifestations. The variability in clinical indicators among patients could result from individual differences or differences among the strains ofB. burgdorferithat initiate the infections. Strains ofB. burgdorferican be classified into subtypes based on various typing methods. Increasing evidence suggests that certain subtypes are more likely to cause hematogenous dissemination than others [56]. Those facts that BmpA can be expressed from three independent transcripts bmpA, bmpAbmpB and bmpCbmpA, and that leader peptide is located in front of bmpA transcript and can regulate bmpA translation, suggest that differences in BmpA expression can be involved in virulence strain diversity.
Data Availability
Data used for this publication is available from the corresponding author upon request.
Conflict of interest
There are no conflicts of interest.
Acknowledgment
Autor is thankful to Dr. Cabello for opportunity to perform and publish this research as an independent research project.
There are no references