Journal Name: Journal of Biomedical Research and Reviews
Article Type: Research
Received date: 31-May-2022
Accepted date: 23-June-2022
Published date: 30-June-2022
Citation: Mohammadisoleimani E, Firoozi Z, Haghi-Aminjan H, Naghizadeh MM, Zeighami S et al. (2022) PGGHG and ODF3B as New Tumor Suppressor Genes in Renal Cell Carcinoma. J Biomed Res Rev Vol: 5, Issu: 1. (44-53).
Copyright: © 2022 Mohammadisoleimani E et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Renal cell carcinoma (RCC) is the most frequent heterogeneous type of kidney cancer. Although the role of many genes in the development of RCC has been shown, its specific molecular mechanism is still unknown.
Objective: In this research, we studied the expression profile of two uncharacterized genes, PGGHG and ODF3B, with possible role in renal cancers in our RCC patients.
Methods: Using TCGA data, firstly, we looked for significant downregulated genes in RCC with poor overall survival (OS) time. Then, we only considered two uncharacterized genes with involvement in the same protein network in this cancer including ODF3B and PGGHG. Their expression was determined using the QPCR in our 40 RCC patients. Moreover, Enrichr was used to investigate their pathways, ontologies, and possible upstream transcription factors.
Results:In contract to TCGA, our study revealed the low expression of PGGHG and ODF3B in our KIRC patients. The expression level of ODF3B in the patients who had tumor size > 4cm was significant. Our data found the possible upstream transcription factor of ODF3B and PGGHG regulating their expression in biological pathways, mainly PPARG as the same upstream transcription factor affecting both genes.
Conclusion: Since our study showed lower expression of these genes in our patients in contrast to TCGA data, more studies from different population with higher number of patients are needed to find whether different population and statuses are involved in this reverse result and also to determine the precise mechanism of their involvement in RCC pathogenesis.
Keywords
Carcinoma, ODF3B, PGGHG, Renal Cell.
Abstract
Background: Renal cell carcinoma (RCC) is the most frequent heterogeneous type of kidney cancer. Although the role of many genes in the development of RCC has been shown, its specific molecular mechanism is still unknown.
Objective: In this research, we studied the expression profile of two uncharacterized genes, PGGHG and ODF3B, with possible role in renal cancers in our RCC patients.
Methods: Using TCGA data, firstly, we looked for significant downregulated genes in RCC with poor overall survival (OS) time. Then, we only considered two uncharacterized genes with involvement in the same protein network in this cancer including ODF3B and PGGHG. Their expression was determined using the QPCR in our 40 RCC patients. Moreover, Enrichr was used to investigate their pathways, ontologies, and possible upstream transcription factors.
Results:In contract to TCGA, our study revealed the low expression of PGGHG and ODF3B in our KIRC patients. The expression level of ODF3B in the patients who had tumor size > 4cm was significant. Our data found the possible upstream transcription factor of ODF3B and PGGHG regulating their expression in biological pathways, mainly PPARG as the same upstream transcription factor affecting both genes.
Conclusion: Since our study showed lower expression of these genes in our patients in contrast to TCGA data, more studies from different population with higher number of patients are needed to find whether different population and statuses are involved in this reverse result and also to determine the precise mechanism of their involvement in RCC pathogenesis.
Keywords
Carcinoma, ODF3B, PGGHG, Renal Cell.
Introduction
Renal cell carcinoma (RCC) is the most common type of kidney malignancies and highly aggressive heterogeneous disease [1,2]. The most common histological subtypes of RCC are clear cell, papillary, and chromophobe responsible for 70%, 10–15%, and 5% of the RCC types, respectively [3,4]. So far, many factors have been implicated in the development of ccRCC, including genetic and environmental factors. Moreover, dysregulation of several genes has been contributed to the RCC progression and metastasis in different pathways. Clear cell RCC patients respond poorly to radiotherapy and chemotherapy. Therefore, the molecular predictions of disease progression and metastasis are important for therapies. We are looking to explore new molecular biomarkers of disease progression and ccRCCspecific molecular mechanisms that may provide new targeted treatment options.
TCGA data from GDC Data Portal (https://portal. gdc.cancer.gov) contains a huge genomic, epigenomic, transcriptomic, and proteomic data of numerous cancer types which can help scientists to use its data for investigation of uncharacterized genes in cancers. Using analysis of TCGA renal cancers data, we found altered expression of two uncharacterized genes, including proteinglucosylgalactosylhydroxylysine glucosidase (PGGHG) (also known as ATHL1) and outer the dense fiber of sperm tails 3B (ODF3B) (Also known as FAP123; ODF3L3) in KIRC. Their altered expression was correlated with worse prognosis with reduced survival in the KIRC patients. Moreover, using STRING database we found their involvement in the same network. Therefore, our study set out to investigate the potential contribution of these two genes with unknown roles, in RCC pathogenesis.
The chromosomal positions of ODF3B is 22q13.33. The pronephros is consisted of two types of epithelial cells, including transportive and multiciliated cells (MCCs). The epithelial MCCs expresses ODF3B protein [5]. Previous study showed that ODF3B transcripts localize in pronephros cells and labels maturing MCCs in renal progenitors [6].
In relation to the PGGHG (located on 11p15.5), it was found to act in release of glucose from human placental type IV collagen [7]. Data extracted from Enrichr (https:// maayanlab.cloud/Enrichr/) shows its involvement in hydrolase activity, hydrolyzing O-glycosyl compounds (GO: 0004553). Moreover, previous study conducted by Wei Shi et al. showed higher expression of hsa-mir-484 correlated with worse prognosis in breast cancer. In their study, they found that one of target gene of hsa-mir-484 was PGGHG [8]. Based on these evidence, identification of the role of these two genes in renal cancer may help find new pathways in this cancer.
Materials and MAethods
Gene selection
Using UALCAN webserver (http://ualcan.path.uab. edu.), firstly, we looked for significant upregulated genes in TCGA-KIRC. Next, we looked for genes with poor overall survival (OS) time with worse prognosis among them, and then we only considered two uncharacterized genes in KIRC including ODF3B and PGGHG. Moreover, we investigated the expression of these genes in other two types of renal cancer including Kidney renal papillary cell carcinoma (KIRP) and Kidney chromophobe (KICH). In addition to these data, to strengthen their role in renal cancers, we looked for their protein network using STRING (https://string-db.org) to find whether they are connected in same protein network and also to help identify their possible pathways.
Patient characteristics and tumor samples
To investigate mRNA expression of PGGHG and ODF3B in renal cancers, we collected 40 tumor tissues and their adjacent normal tissues of RCC patients from Ali-Asghar, Namazi, and Ghadir Mother and Child Hospitals (Shiraz, Iran). In total, RCC patients had not received radiotherapy and chemotherapy before surgery. The samples were immediately immersed in liquid nitrogen and stored at -80°C until use. The clinicopathologic and demographic features of RCC patients are shown in Table 1. After surgery, we followed up patients to find they had all-cause mortality.
RNA extraction and cDNA synthesis
Using the Trizol isolation reagent (Invitrogen, Thermo Fisher) total RNA was extracted from each sample according to the manufacturer’s instructions. Gel electrophoresis and a nanodrop spectrophotometer (BioTek, HTX multimode reader) was used to assess RNA quality and quantity, respectively. Total RNA was reverse-transcribed into firststrand cDNA using the PrimeScript™ RT Reagent Kit (Takara, Cat.No: RR037A).
qRT-PCR
QRT-PCR was carried out using Power SYBR® Green PCR Master Mix (ABI, USA) on the 7500 real-time PCR system (ABI, life technology). 7.5 μl BioFACT™ master mix including SYBR Green (Ampliqon, Cat.No: A325402-25), along with 1 μl of cDNA, 0.75 μl of each primer and 5 μl DNase-free dH2O was used for total volume of 15 μl reaction mix. B2M was used as an internal control. The sequences for primers are shown in Supplementary Table 1. Relative quantification was calculated by the 2-ΔΔCT.
Enrichment analysis
In our study we also used Enrichr database (https:// maayanlab.cloud/Enrichr/) to look for their pathways, ontologies, and possible upstream transcription factors and among others. To evaluate survival analysis our patients, we used the Kaplan–Meier method based on observed survival times. This analysis was performed using IBM SPSS version 26 software (IBM Corp, Armonk, NY).
Statistical analysis
Statistical analyses were done in IBM SPSS version 26 software (IBM Corp, Armonk, NY). The data were presented as mean and standard deviation for ΔΔCt, or median for fold change. Wilcoxon test was used to compare fold changes between tumors and adjacent normal renal tissues.
Table 1: The association of ODF3B and PGGHG expression levels with demographic and clinicopathological factors in RCC patients.
| ODF3B level | PGGHG level | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Mean | SD | Median | P-value | N | Mean | SD | Median | P-value | ||
| Sex | male | 29 | 0.924 | 0.984 | 0.620 | 0.844 | 29 | 0.795 | 1.037 | 0.309 | 0.296 |
| female | 11 | 0.748 | 0.637 | 0.660 | 11 | 0.971 | 0.857 | 0.798 | |||
| Tumor size | ≤4 | 20 | 1.054 | 0.881 | 0.919 | 0.060 | 20 | 0.758 | 0.843 | 0.540 | 0.745 |
| >4 | 20 | 0.697 | 0.899 | 0.357 | 20 | 0.928 | 1.122 | 0.333 | |||
| Tumor focality | focal | 25 | 1.097 | 1.031 | 0.774 | 0.067 | 25 | 0.840 | 1.039 | 0.491 | 0.856 |
| unifocal | 15 | 0.507 | 0.434 | 0.587 | 15 | 0.849 | 0.918 | 0.309 | |||
| Tumor type | clear cell | 27 | 0.959 | 0.835 | 0.688 | 0.004 | 27 | 0.925 | 0.934 | 0.505 | 0.017 |
| papillary | 5 | 1.283 | 1.330 | 0.774 | 5 | 0.974 | 0.958 | 0.644 | |||
| chromophore | 6 | 0.043 | 0.030 | 0.031 | 6 | 0.069 | 0.096 | 0.029 | |||
| oncocytoma | 2 | 1.231 | 0.864 | 1.231 | 2 | 1.730 | 2.427 | 1.730 | |||
| Tumor necrosis | seen | 20 | 0.924 | 1.086 | 0.601 | 0.715 | 20 | 0.598 | 0.771 | 0.331 | 0.160 |
| not seen | 20 | 0.827 | 0.685 | 0.674 | 20 | 1.088 | 1.124 | 0.635 | |||
| Fuhrman nuclear grade | 1 | 5 | 0.328 | 0.308 | 0.233 | 0.113 | 5 | 0.359 | 0.573 | 0.090 | 0.225 |
| 2 | 21 | 0.768 | 0.920 | 0.470 | 21 | 0.818 | 0.961 | 0.399 | |||
| 3 | 9 | 1.189 | 0.711 | 1.603 | 9 | 1.268 | 1.180 | 0.798 | |||
| 4 | 5 | 1.310 | 1.271 | 1.065 | 5 | 0.665 | 0.979 | 0.302 | |||
| Lymph vascular perineural invasion | No | 8 | 1.057 | 1.055 | 0.841 | 0.467 | 8 | 0.801 | 1.042 | 0.304 | 0.866 |
| Yes | 32 | 0.830 | 0.866 | 0.625 | 32 | 0.854 | 0.985 | 0.445 | |||
| Extension | No | 30 | 0.870 | 0.884 | 0.625 | 1.000 | 30 | 0.901 | 1.046 | 0.445 | 0.779 |
| Yes | 10 | 0.892 | 0.985 | 0.695 | 10 | 0.670 | 0.787 | 0.330 | |||
| Cancer history | No | 19 | 0.891 | 0.998 | 0.587 | 0.882 | 19 | 0.897 | 1.085 | 0.399 | 0.903 |
| Yes | 21 | 0.861 | 0.820 | 0.774 | 21 | 0.794 | 0.906 | 0.333 | |||
| BMI | ≤25 | 9 | 1.224 | 1.433 | 0.616 | 0.918 | 9 | 0.762 | 0.928 | 0.234 | 0.784 |
| 25-29 | 26 | 0.771 | 0.689 | 0.645 | 26 | 0.826 | 1.007 | 0.445 | |||
| ≥30 | 5 | 0.791 | 0.650 | 0.620 | 5 | 1.080 | 1.134 | 0.798 | |||
| Kidney disease | No | 29 | 0.867 | 0.838 | 0.629 | 0.880 | 29 | 0.880 | 1.038 | 0.357 | 0.940 |
| Yes | 11 | 0.897 | 1.083 | 0.470 | 11 | 0.745 | 0.860 | 0.399 | |||
Table 2: The association of ODF3B and PGGHG expression levels with demographic and clinicopathological factors in RCC patients, according to categorizing patients in to two groups of high and low expressions.
| ODF3B level | PGGHG level | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | P-value | Low | High | P-value | ||||||
| N | % | N | % | N | % | N | % | ||||
| Tumor size | ≤4 | 6 | 30.0% | 14 | 70.0% | 0.011 | 9 | 45.0% | 11 | 55.0% | 0.527 |
| >4 | 14 | 70.0% | 6 | 30.0% | 11 | 55.0% | 9 | 45.0% | |||
| Tumor focality | focal | 11 | 44.0% | 14 | 56.0% | 0.327 | 12 | 48.0% | 13 | 52.0% | 0.744 |
| unifocal | 9 | 60.0% | 6 | 40.0% | 8 | 53.3% | 7 | 46.7% | |||
| Fuhrman nuclear grade | 1 | 4 | 80.0% | 1 | 20.0% | 0.157 | 4 | 80.0% | 1 | 20.0% | 0.093 |
| 2 | 12 | 57.1% | 9 | 42.9% | 10 | 47.6% | 11 | 52.4% | |||
| 3 | 2 | 22.2% | 7 | 77.8% | 2 | 22.2% | 7 | 77.8% | |||
| 4 | 2 | 40.0% | 3 | 60.0% | 4 | 80.0% | 1 | 20.0% | |||
| Lymph vascular perineural invasion | No | 4 | 50.0% | 4 | 50.0% | 1.000 | 5 | 62.5% | 3 | 37.5% | 0.429 |
| Yes | 16 | 50.0% | 16 | 50.0% | 15 | 46.9% | 17 | 53.1% | |||
| BMI | ≤25 | 5 | 55.6% | 4 | 44.4% | 0.793 | 6 | 66.7% | 3 | 33.3% | 0.508 |
| 25-29 | 12 | 46.2% | 14 | 53.8% | 12 | 46.2% | 14 | 53.8% | |||
| ≥30 | 3 | 60.0% | 2 | 40.0% | 2 | 40.0% | 3 | 60.0% | |||
| Background disease | None | 11 | 52.4% | 10 | 47.6% | 0.727 | 12 | 57.1% | 9 | 42.9% | 0.313 |
| High blood pressure | 7 | 58.3% | 5 | 41.7% | 6 | 50.0% | 6 | 50.0% | |||
| diabetes | 1 | 33.3% | 2 | 66.7% | 1 | 33.3% | 2 | 66.7% | |||
| High blood pressure and diabetes | 1 | 33.3% | 2 | 66.7% | 0 | 0.0% | 3 | 100.0% | |||
| prostate problems | 0 | 0.0% | 1 | 100.0% | 1 | 100.0% | 0 | 0.0% | |||
| Kidney disease | No | 14 | 48.3% | 15 | 51.7% | 0.723 | 15 | 51.7% | 14 | 48.3% | 0.723 |
| Yes | 6 | 54.5% | 5 | 45.5% | 5 | 45.5% | 6 | 54.5% | |||
| Kidney stone | No | 10 | 47.6% | 11 | 52.4% | 0.752 | 10 | 47.6% | 11 | 52.4% | 0.752 |
| Yes | 10 | 52.6% | 9 | 47.4% | 10 | 52.6% | 9 | 47.4% | |||
The relation of PGGHG and ODF3B expression with demographic and clinicopathological features was assessed by Mann–Whitney and Kruskal-Wallis tests. In the next step, fold changes were divided into 2 groups of high and low expressions conforming to median for each gene, and the comparison among these groups were analyzed by chi-square test and independent t test. After surgery, we followed up patients until all-cause death used and censured alive one. The log rank test is a statistical hypothesis test that may be used to compare two high and low gene expression survival curves.
Results
TCGA data analysis
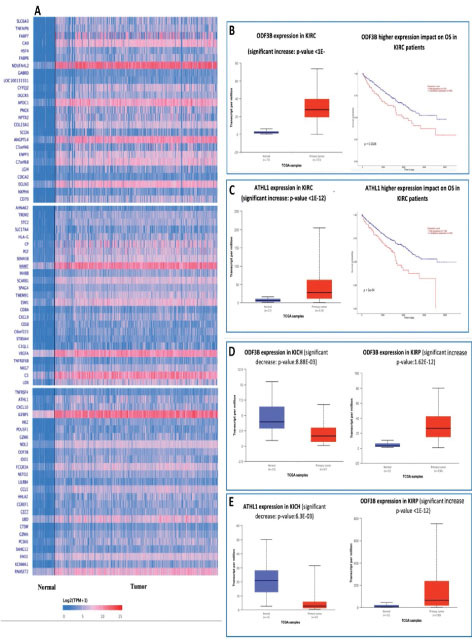
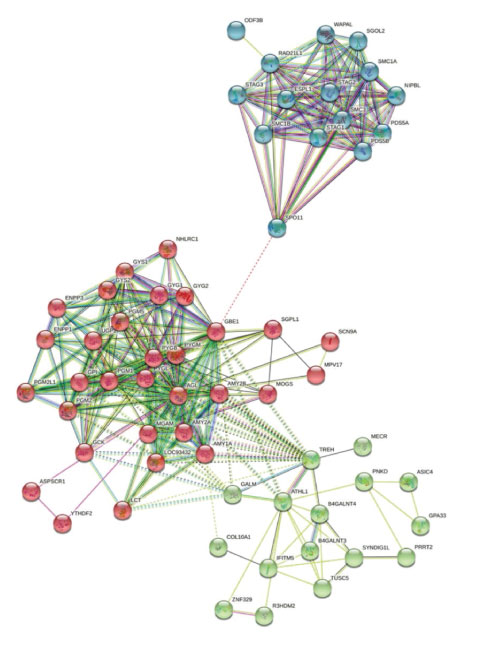
Using TCGA data we extracted 75 top upregulated genes in KIRC shown as heatmap in Figure 1A (extracted from UALCAN webserver (http://ualcan.path.uab.edu.), a heatmap containing some of these genes with similar patterns have been reported by Ping Wu et al. [9]. Then among theme, we selected only two uncharacterized genes with worse prognosis with reduced survival in KIRC (Figure 1AC). Moreover, we investigated their expression in other two renal cancer types KIRP and KICH, which both genes showed lower-expression in KICH but over-expression in KIRP similar to the KIRC (Figure 1D and 1E). We also found that these two proteins can have indirect connections in the same protein network (Figure 2, using STRING), suggesting their possible roles in renal pathways and cancers. In addition to these data, its protein network revealed its interaction with several important proteins involved in biological pathways and cancers (including renal cancers), for instance, COL10A1 [10] (Figure 2). Based on these primary data we conducted our experimental study to identify the expression of these two genes in our patients with renal cancers.
Downregulation of PGGHG and ODF3B gene expression in RCC
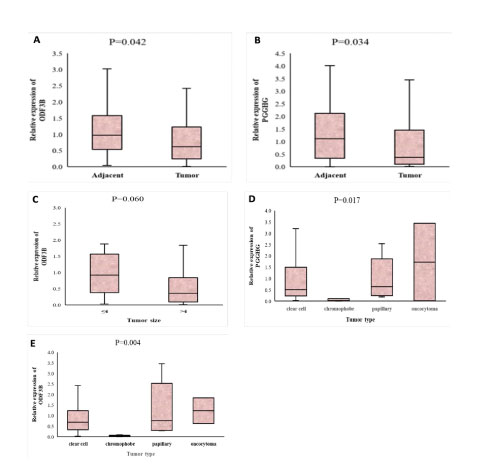
Using qRT-PCR, the expression levels of mRNAs were assessed in 40 tumor samples and their tumor’s adjacent normal tissues. As shown in the figure 3A, the expression level of ODF3B was significantly lower in tumors tissues (median=0.625) in comparison to the tumor’s adjacent normal tissues (median=0.977) (P value=0.042). Also, the expression of PGGHG had significantly lower in tumor tissues (median=0.378) compared to their adjacent normal tissues (median=1.120) (Pvalue=0.034, Figure 3B).
Association between PGGHG and ODF3B gene expression and clinicopathological and demographic features of RCC
Next, we investigated the relationship between PGGHG and ODF3B expression levels and the clinicopathologic status of patients with renal cell carcinoma. Our results demonstrated that ODF3B had lower expression in RCC patients who had tumor size>4cm (P value=0.060, Figure 3C). When we divided fold changes into two high and low expressions groups, chi-square test proved the significant relation between tumor size and ODF3B expression (P value= 0.011, Table 2). Comparison of PGGHG expression level and tumor size did not show a significant difference (P value=0.744).
Since ccRCC is the most common of RCC and oncocytoma is the least common type in our study community (Table 1), our analysis showed that PGGHG expression level was higher in oncocytoma type in comparison to other type of RCC (P value=0.017, Figure 3D). Similarly, the relationship between the ODF3B expression level and the tumor type showed significant. The expression level of ODF3B was higher in oncocytoma type (P value=0.004, Figure 3E).
Functional enrichment analysis
Using Enrichr, we found down-regulation of ODF3B upon RUNX1 Knock-out mouse, PPARG deficiency mouse, SETDB1 Knock-down in THP1 human cells, IRF9 Knock-down in human cells, NANOG over-expressed mouse, and CREM Knock-out mouse (Supplementary Table 2). These data can suggest the possible upstream transcription factors of ODF3B which regulate its expression in biological pathways. Among GO ontologies using Enrichr, ODF3B was found to be involved in cytoskeleton (GO: 0005856, p-value: 0.03000) in terms of cellular component (CC).
Figure 1: A. Seventy-five top upregulated genes in TCGA-KIRC from UALCAN webserver. B. Expression of ODF3B in KIRC and effect of its altered expression on OS in TCGA-KIRC. C. Expression of PGGHG in KIRC and effect of its altered expression on OS in TCGA-KIRC. D. Expression of ODF3B in KICH and KIRP. E. Expression of PGGHG in KICH and KIRP.
Figure 2: Protein network of PGGHG and ODF3B. It shows connection between protein networks of PGGHG and ODF3B, representing their possible roles in the same pathways and cancer pathogenesis.
Figure 3: A and B. The boxplot of comparison of PGGHG and ODF3B expression between tumor samples and their paired adjacent normal tissues, respectively. C. The association between the expression of ODF3B and tumor size D. PGGHG expression levels in the different subgroups of tumor types in RCC. E. ODF3B expression levels in the different subgroups of tumor types in RCC.
In relation to the PGGHG, Enrichr revealed its downregulation upon ZNF503 shRNA in H1 human cells, NFYC Knock-out in mouse, BCL6 Knock-down in human cells, POU2AF1 over-expression in mouse, EPAS1 Knock-down in HUVEC human cells, NFYA Knock-out in mouse, FOXP3 activation in human cells, PPARG over-expression in mouse, and GATA4 over-expression in A549 human cells (Supplementary Table 3). The interesting result was that while PPARG over-expression in mouse resulted in PGGHG down-regulation, PPARG deficiency mouse showed ODF3B down-regulation. These data may indicate PPARG as the same upstream transcription factor affecting ODF3B and PGGHG. Among GO using Enrichr, PGGHG showed involvement in hydrolase activity, hydrolyzing O-glycosyl compounds (GO:0004553, p-value: 0.001850, in terms of molecular function (MF).
As shown in Figure 2, protein networks of PGGHG and ODF3B are connected and correlated. Therefore, we investigated the GO terms for all proteins involved in these networks to shed light on the identification of possible pathways of PGGHG and ODF3B.
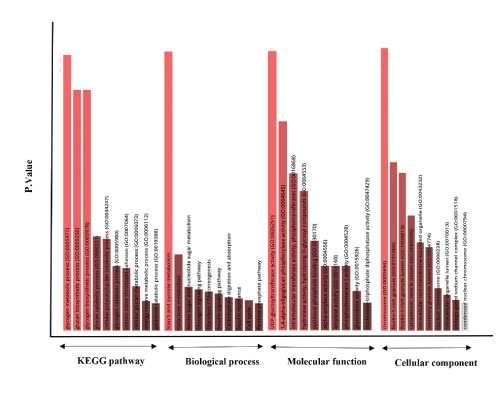
Enrichr revealed significant KEGG, BP, MF, and CC for proteins involved in these networks given in Figure 4. However, the highest significant terms were as follow: KEGG: Starch and sucrose metabolism (p-value: 3.058e-43); BP: glucan biosynthetic process (GO:0009250, p-value: 5.259e- 24), glycogen biosynthetic process (GO:0005978, p-value: 5.259e-24), and glycogen metabolic process (GO:0005977, p-value: 8.723e-27); MF: UDP-glucosyltransferase activity (GO:0035251, p-value: 1.319e-8) and 1,4-alpha-oligoglucan phosphorylase activity (GO:0004645, p-value: 2.305e- 7); CC: chromosome (GO:0005694, p-value: 1.921e-8). All information and proteins involved in these terms are provided in Supplementary file 1. These data may propose the possible roles of PGGHG and ODF3B, directly or indirectly, in these GO terms and KEGG pathways.
Figure 4: Significant KEGG (Kyoto Encyclopedia of Genes and Genomes) human pathways, BP (biological process), MF (molecular function), and CC (cellular component) GO terms for proteins involved in PGGHG and ODF3B networks.
Overall survival
To evaluate survival analysis, we used the Kaplan–Meier method based on observed survival times. However, we were not able to predict the prognosis of renal cell carcinoma patients based on PGGHG and ODF3B gene expression using this data (Supplementary figure 1). The small proportion of participants evaluated, low duration of follow up and undetected exact cause of death could be some of the reasons.
Discussion
Renal cell carcinoma (RCC) is the most common form of kidney cancer and accounts for approximately 3.7% of the total cancer deaths [11]. Identification of biomarkers helps in improving the prognosis and early detection of RCC. Despite astonishing molecular advances and their significant impact on the detection of functional mRNAs in malignancies, numerous questions remain regarding molecular interactions and their function and need further attention.
In this study, we compared the PGGHG and ODF3B expression levels in tumor and their adjacent normal tissues in RCC patients and their association with demographical and clinicopathologic factors.
Our study with the use of QPCR as gold standard for relative expression analysis revealed the downregulated level of PGGHG and ODF3B in the tumor tissues compared to their adjacent normal tissues which were in contract to TCGA-KIRC. The reverse data for both genes in our experiments can strengthen the involvement of both proteinss in the same network. There are some reasons which might show these reverse results as follow: 1- The TCGA data is the raw data of RNA sequencing which usually are validated by gold standard approaches such as QPCR. Since in some genes the coverage is not optimal and may affect the correct overall gene expression. 2- Another factor which might affect the results is the population diversity since the TCGA data only has 8 KIRC samples from Asian region and we don’t know how many samples are from Iranian population. 3- Other environmental factors and life styles in different area might change the expression of these genes. However, since our study showed lower expression of these genes in all of our patients which was in contrast to TCGA data, more researches from different population with higher number of patients are needed to find whether different population, statuses, and other factors are involved in this reverse result, helping determination of the precise mechanism of their involvement in RCC pathogenesis.
Our results indicate that the expression level of ODF3B is significantly lower in tumor tissues than their normal adjacent ones. There are few investigations about the role of the studied genes in the development and progression of cancers. In the case of the ODF3B gene, an experimental study related to cancer has not been reported. Jihye Ryu et al. examine promotor activity of ODF3B in multiple sclerosis (MS) on large scale by genome-wide association studies (GWAS) signals. In this study, an association of promotor variants with the expression of this gene was reported [12]. In another study, Honglin Zhu et al. showed the ODF3B as potential methylation-regulated differentially expressed gene [13]. This gene delineates the abnormal activation of immune regulation in the pathogenesis of Systemic sclerosis (SSc).
Our study also showed significant lower expression of PGGHG in RCC tissues in comparison to their adjacent normal specimens. In the past decades, the role of aerobic glycolysis in cancer cells has been discussed, however, data supporting for glycogen biology in RCC are lacking [14]. In line with our results, a study demonstrated that reduced AGL, a glycogen debranching enzyme, increased tumor growth in patients with bladder cancer by RNA interference screen [15]. In another study, AGL loss led to the progression of bladder cancer. Details of this metabolomics pathway showed increased glucose metabolism with the help of serine hydroxymethyltransferase in cells [16].
Our data from Enrichr revealed significant KEGG, MF, BP, and CC for proteins involved in ODF3B and PGGHG networks, suggesting the possible roles of these two proteins, directly or indirectly, in these GO terms and KEGG pathways. Moreover, using Enrichr, we found the downregulation of ODF3B and PGGHG upon perturbations of some transcription factors, mainly PPARG which affected both genes. PPARG, Peroxisome Proliferator Activated Receptor Gamma, is involved in adipocyte differentiation considered a “master regulator” of adipogenesis. Moreover, it represses inflammatory response genes in mouse macrophages [17,18]. We also identified the positive relationship of tumor size with the ODF3B expression level (Table 2). According to a study by Zhi et al., an increase in tumor size has been reported in the progression and high risk of lymph node metastases (LNM) in ccRCC [19].
Conclusion
In summary, we found lower expression of ODF3B and PGGHG in renal cancers and proposed their possible biological pathways and upstream transcription factors. Our results propose investigation of functional studies for ODF3B and PGGHG genes to identify their exact roles in biological pathways and cancers. While our study with the use of QPCR showed the lower-expression of PGGHG and ODF3B in our patients, they were in contract to TCGA-KIRC and some above-mentioned reasons might answer this reverse result. However, our study showed lower expression of these genes in all of our patients and further studies from different population with higher number of patients are needed to investigate the expression of these genes in renal cancers to uncover the exact role of them in these cancers.
Authors’ Contributions
Conceptualization: [Elham Mohammadisoleimani]; Methodology: [Elham Mohammadisoleimani, Zahra Firoozi, and Hassan Dastsooz]; Formal analysis and investigation: [Hassan Dastsooz , Mohammad Mehdi Naghizadeh, and Elham Mohammadisoleimani]; Writing - original draft preparation: [Elham Mohammadisoleimani, Hamed Haghi- Aminjan, Hassan Dastsooz, and Zahra Firoozi]; Writing - review and editing: [Hassan Dastsooz, Abdolreza Daraei, and Yaser Mansoori]; Funding acquisition: [Yaser Mansoori], Resources: [Shahryar Zeighami, Ali Ariafar, Hosein Mansoori]; Supervision: [Yaser Mansoori]. All authors read and approved the manuscript before submission.
Ethical Statements
The local ethical committee at Fasa University of Medical Sciences (ethical code: IR.FUMS.REC.1398.147) approved this study.
Conflict of interest
None. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The written informed consent was obtained from all participants. Samples were provided from Ali-Asghar, Namazi, and Ghadir Mother and Child hospitals, Shiraz, Iran. The authors are thankful to patients who took part in this study. This study was supported by Fasa University of Medical Sciences (FUMS), Fasa, Iran (Grant Number: 97423).
Wei W, Lv Y, Gan Z, Zhang Y, Han X, et al. (2019) Identification of key genes involved in the metastasis of clear cell renal cell carcinoma. Oncol Lett 17: 4321-4328. [ Ref ]
Scoll BJ, Wong YN, Egleston BL, Kunkle DA, Saad IR, et al. (2009) Age, tumor size and relative survival of patients with localized renal cell carcinoma: a surveillance, epidemiology and end results analysis. J Urol 181: 506-511. [ Ref ]
Muglia VF, Prando A (2015) Renal cell carcinoma: histological classification and correlation with imaging findings. Radiol Bras 48: 166-174. [ Ref ]
Znaor A, Lortet-Tieulent J, Laversanne M, Jemal A, Bray F (2015) International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol 67: 519-530. [ Ref ]
Marra AN, Adeeb BD, Chambers BE, Drummond BE, Ulrich M, et al. (2019) Prostaglandin signaling regulates renal multiciliated cell specification and maturation. Proceedings of the National Academy of Sciences of the United States of America 116: 8409-8418. [ Ref ]
Marra AN, Cheng CN, Adeeb B, Addiego A, Wesselman HM, et al. (2019) Iroquois transcription factor irx2a is required for multiciliated and transporter cell fate decisions during zebrafish pronephros development. Scientific Reports 9: 6454. [ Ref ]
Hamazaki H, Hamazaki MH (2016) Catalytic site of human proteinglucosylgalactosylhydroxylysine glucosidase: Three crucial carboxyl residues were determined by cloning and site-directed mutagenesis. Biochem Biophys Res Commun 469: 357-362. [ Ref ]
Shi W, Dong F, Jiang Y, Lu L, Wang C, et al. (2019) Construction of prognostic microRNA signature for human invasive breast cancer by integrated analysis. Onco Targets Ther 12: 1979-2010. [ Ref ]
Wu P, Xiang T, Wang J, Lv R, Wu G (2020) TYROBP is a potential prognostic biomarker of clear cell renal cell carcinoma. FEBS Open Bio 10: 2588-604. [ Ref ]
Chen S, Wei Y, Liu H, Gong Y, Zhou Y, et al. (2021) Analysis of Collagen type X alpha 1 (COL10A1) expression and prognostic significance in gastric cancer based on bioinformatics. Bioengineered 12: 127-137. [ Ref ]
Chen YY, Hu HH, Wang YN, Liu JR, Liu HJ, et al. (2020) Metabolomics in renal cell carcinoma: From biomarker identification to pathomechanism insights. Arch Biochem Biophys 695: 108623. [ Ref ]
Ryu J, Woo J, Shin J, Ryoo H, Kim Y, et al. (2014) Profile of differential promoter activity by nucleotide substitution at GWAS signals for multiple sclerosis. Medicine (Baltimore) 93: e281. [ Ref ]
Zhu H, Zhu C, Mi W, Chen T, Zhao H, et al. (2018) Integration of Genome-Wide DNA Methylation and Transcription Uncovered Aberrant Methylation-Regulated Genes and Pathways in the Peripheral Blood Mononuclear Cells of Systemic Sclerosis. Int J Rheumatol 2018: 7342472. [ Ref ]
Xie H, Song J, Godfrey J, Riscal R, Skuli N, et al. (2021) Glycogen metabolism is dispensable for tumour progression in clear cell renal cell carcinoma. Nat Metab 3: 327-336. [ Ref ]
Guin S, Pollard C, Ru Y, Ritterson Lew C, Duex JE, et al. (2014) Role in tumor growth of a glycogen debranching enzyme lost in glycogen storage disease. J Natl Cancer Inst 106: dju062. [ Ref ]
Weinhaus B, Guin S (2017) Involvement of glycogen debranching enzyme in bladder cancer. Biomed Rep 6: 595-598. [ Ref ]
Evans RM, Barish GD, Wang Y-X (2004) PPARs and the complex journey to obesity. Nature Medicine 10: 355-361. [ Ref ]
Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, et al. (2005) A SUMOylationdependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 437: 759-763. [ Ref ]
Zhi Y, Li X, Qi F, Hu X, Xu W (2020) Association of Tumor Size with Risk of Lymph Node Metastasis in Clear Cell Renal Cell Carcinoma: A Population-Based Study. J Oncol 2020: 8887782. [ Ref ]