Journal Name: Journal of Pediatrics and Infants
Article Type:Research Article
Received date:31 December, 2020
Accepted date:03 February, 2021
Published date: 10 February, 2021
Citation:Chen H, Yan H, Chen C, Cao Y, Zhang X (2021) Associations between Apolipoprotein E (APOE) Polymorphisms and Cerebral Palsy: A Meta-Analysis. J Pediat Infants Vol: 4, Issu: 1 (40-48).
Copyright:© 2021 Chen H et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Apolipoprotein E (APOE) is one of the main apolipoproteins that plays an important role in the central neuronal system. The relationship between its polymorphisms and cerebral palsy (CP) is ambiguous. We conducted eligible studies identified from Elsevier Science Direct, PubMed, Springer Link, WEB OF SCIENCE, Chinese National Knowledge Infrastructure and WanFang Data up to February 2019 to conduct a systematic review. In total, 10 eligible studies were included in this meta-analysis (1570 CP patients and 1982 healthy subjects). Significant associations with CP were observed for APOE polymorphisms in allele (ε4: P < 0.001, OR 2.05, 95% CI 1.40 to 2.99; ε2: P = 0.04, OR 1.41, 95% CI 1.01 to 1.96) and dominant (E4 carriers: P = 0.004, OR 1.90, 95% CI 1.23 to 2.92) models in overall analyses. Interestingly, subgroup analysis indicated a significantly increased risk for CP in Chinese individuals with APOE ε4 (P<0.00001, OR 3.71, 95% CI 2.37 to 5.78) and in E4 carriers (P<0.00001, OR 3.95, 95% CI 2.38 to 6.53) but not with in those with APOE ε2 (P=0.69, OR 1.09, 95% CI 0.72 to 1.65). Combined with the results of our analysis, we concluded that the risk of CP was significantly increased in individuals with the ε4 allele. However, meta-analysis yielded an incongruent result for the APOE ε2 allele between multiethnic samples and the Chinese subgroup. These conclusions should be confirmed through further studies.
Keywords: Apolipoprotein E; Cerebral palsy; Meta-analysis; Gene polymorphisms.
Introduction
Cerebral palsy (CP) is a group of motor and posture developmental disorders caused by non-progressive injuries in developing foetuses or infants, resulting in disordered movement and coordination. CP is a severe disability in children, with 40% of affected children being unable to walk independently, 1/3 having epilepsy, up to 1/3 being non-verbal and approximately 1/2 having some degree of cognitive impairment [1- 6]. In recent years, evidence from several high-income countries (United States, Australia, Europe, Canada, Sweden, and Japan) has shown that the prevalence of CP has decreased (mainly in low birth weight and premature infants) but still remains at 2‰ ~ 3‰ [6]. Epidemiological survey results of more than 320,000 children aged 1-6 years old in 12 provinces and autonomous regions of China in 2013 showed that the prevalence of CP was 2.46‰, which was consistent with the international average [7]. In the United States, children with CP are estimated to cost at least $1 million per person for health care, educational needs, social services, and lost economic opportunities [8]. The prevalence, severity, and burden of CP is becoming an important public health problem threatening children’s health.
The pathogenesis of CP is multifactorial and varied and the causes are premature birth and inflammatory, anoxic environmental, traumatic, metabolic and genetic factors. Previous studies on the pathogenesis of CP have focused on the clinical aetiology. In recent years, both domestic and foreign studies have found that genetic factors are also involved in the aetiology of CP, while the apolipoprotein E (APOE) genotype is one of the most studied genetic risk factors. Apolipoprotein E plays an important role in the distribution of lipids in peripheral tissues such as the peripheral nerve, arterial wall, and brain. The role of APOE with relevance to therapeutic development and treatment of Alzheimer’s disease has accelerated in recent years and may now be relevant to CP treatment. The authors findings are now important and combine environmental, metabolic and genetic factors to be closely linked to the induction of CP. In recent research the anti-aging gene Sirtuin 1 has been shown to be linked to various metabolic diseases (obesity, diabetes, NAFLD) and neurodegenerative diseases. The role of therapeutics with relevance to CP treatment may require Sirtuin 1 activators that may improve developmental disorders by the increase in the neuroprotective protein Sirtuin 1. APOE has now been shown to be linked to Sirtuin 1 levels and APOE therapeutics (compound identification) have been shown to increase brain Sirtuin 1 levels [9-11]. The human APOE gene produces three protein subtypes: APOE ε2 (112Cys/158Cys), APOE ε4 (112Arg/158Arg) and wild-type APOE ε3 (112Cys/158Arg), and the six genotypes (E2/2, E2/3, E2/4, E3/3, E3/4, and E4/4) are located on chromosome 19q13.2 [12,13]. The APOE ε4 allele has been reported to be related to Alzheimer’s disease, age-related cognitive decline [14,15].
Currently, whether there is a link between the APOE genotype and the risk of CP has been investigated [16-27]. However, the existing results are conflicting. Some studies have shown that there was a highly significant association between the ε2 [20,22] or ε4 [22,23] alleles and the risk of CP, whereas others have shown no association [18,19,21]. Due to the small number of samples, the complex genetic relationship may not be detected in individual studies. The purpose of this research is to comprehensively evaluate the possible relationship between APOE polymorphisms and CP risk.
Methods
Search strategy
We conducted a systematic study of the research articles published up to February 2019 through Elsevier Science Direct, PubMed, Springer Link, WEB OF SCIENCE, Chinese National Knowledge Infrastructure (CNKI, in Chinese) and WanFang Data (in Chinese). Two authors independently searched the literature using the following keywords: (Apolipoprotein E OR APOE) AND (cerebral palsy OR CP) AND (gene OR polymorphism OR genotype OR variation OR allele). Some of the relevant literature in the review articles was reviewed to identify additional publications. Studies that met our eligibility criteria were included in the metaanalysis.
Inclusion criteria
To be included, studies needed to (a) explain the association between the APOE gene polymorphism and CP and (b) offer enough original data of the allele frequency or genotype distribution; in addition, (c) when the same case and control subjects appeared in multiple articles, the study with the largest number of participants was included. Conference reports or summaries were not included.
Data extraction and quality assessment
Two authors (C-HY, Y-H) identified eligible articles independently in accordance with the inclusion criteria. The authors also looked up the following data independently: year of publication, first author’s family name, population, study type, types of CP, gene genotyping methods, source of controls (hospital-based vs. population-based), APOE genotype and allele distribution. The Newcastle Ottawa Scale (NOS) was used to assess the quality of the studies included in the meta-analysis. The genotype distribution reported in percentages was calculated for figures. The Hardy–Weinberg equilibrium (HWE) was evaluated in the control groups by the chi-square test (p<0.05 was considered significant). Extracted data were contrasted; if there were discrepancies, they would be resolved through discussion with the third author (Z-XW).
Data extraction and quality assessment
Two authors (C-HY, Y-H) identified eligible articles independently in accordance with the inclusion criteria. The authors also looked up the following data independently: year of publication, first author’s family name, population, study type, types of CP, gene genotyping methods, source of controls (hospital-based vs. population-based), APOE genotype and allele distribution. The Newcastle Ottawa Scale (NOS) was used to assess the quality of the studies included in the meta-analysis. The genotype distribution reported in percentages was calculated for figures. The Hardy–Weinberg equilibrium (HWE) was evaluated in the control groups by the chi-square test (p<0.05 was considered significant). Extracted data were contrasted; if there were discrepancies, they would be resolved through discussion with the third author (Z-XW).
Meta-analysis methods and bias testing
Based on the allele and genotype frequency between the case and the control, the odds ratio (OR) was adopted to evaluate the intensity of the correlation between the APOE polymorphism and CP susceptibility. We calculated ORs and 95% CIs to assess potential associations between APOE polymorphisms and CP in allele, dominant and recessive models based on genotypic distributions of investigated polymorphisms. The Chinese subgroup was then divided according to ethnicity. On the basis of the Q-test, we used the χ² test to analyse the heterogeneity, which was thought to be statistically significant at a P value <0.05 [28]. To quantify heterogeneity, the I² value was calculated and clarified as follows: no heterogeneity, I² =0%; low heterogeneity, I² =25%, moderate heterogeneity, I² =50% and high heterogeneity, I² =75% [29,30].The summary OR was derived by using the Mantel-Haenszel (MH) method with the assumptions of a fixed effects model, as well as by using the DerSimonian and Laird method with the assumptions of a random-effects model [31,32]. The value of the OR was also evaluated using the Z test, and a P value <0.05 was considered statistically significant.
Publication bias was evaluated by visual examination of Begg’s funnel plots. An asymmetric funnel indicated a publication bias, and after that, Egger’s test was perfomed [33,34]. We have also implemented the Duval and Tweedie nonparametric “trim and fill” process to evaluate the possible impact of publication bias in our meta-analysis [35]. The whole statistical analysis was conducted in Stata 12.0 (Stata Corp, College Station, TX, USA) and RevMan V.5.3 (Cochrane, Oxford, UK).
Results
Description of studies
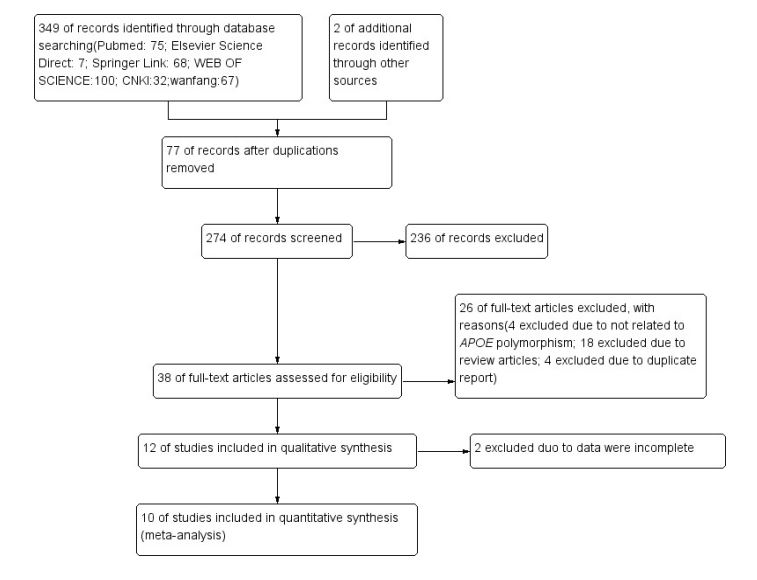
Our literature search generated 351 studies, 274 of which remained when 77 duplications were removed. This number was reduced to 38 after screening the title and abstract (Figure 1). After reading the full text of these papers, 18 studies were excluded, as they were review articles, and another 8 studies were excluded because an overlapping population was analysed or the data were not related to the APOE polymorphism. Then, 12 studies were included in the meta-analysis, but two studies were removed because the data were incomplete. Finally, 10 eligible studies were identified, published from 1995 to 2019, that reported on genotypes of APOE and risk of CP, of which four were published in Chinese [24-27] and the other six were published in English [16-23].
Some studies have been put forward in this field in Brazil, China, the United States, Norway, Australia and Turkey. The combined participants included 1570 CP patients and 1982 healthy subjects. The main features of the studies involved in the meta-analysis are provided in Table 1. We used the NOS rating scale to assess the quality score of each study, as shown in Table 1. The data for the frequencies of APOE alleles and genotypes in the individual studies are shown in Table 1S. The deviation from HWE in the control population was found in three studies [17,21,22].
Overall analyses of the association between APOE polymorphisms and CP susceptibility
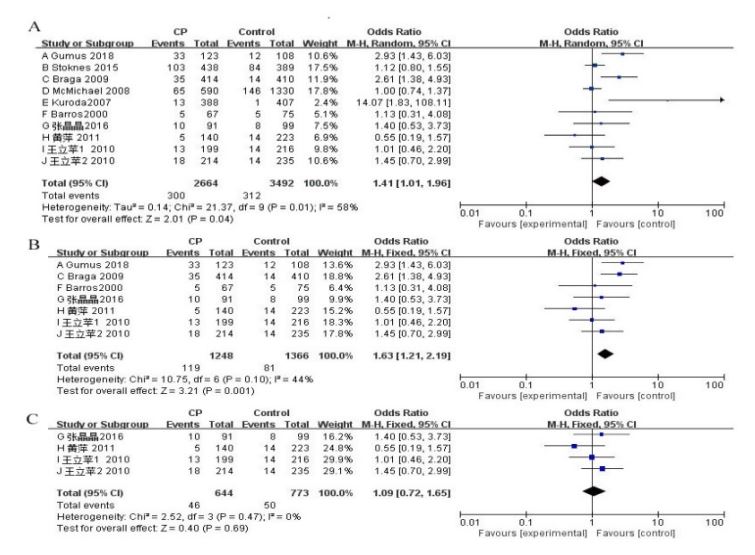
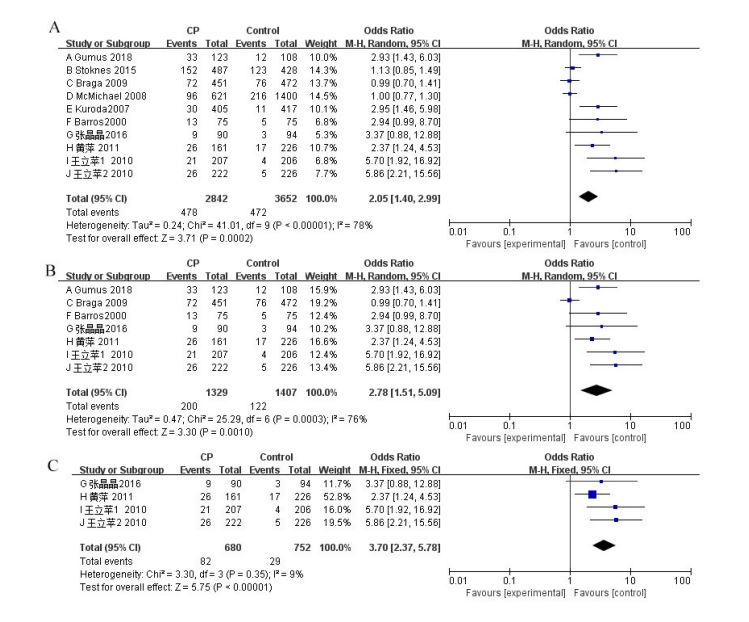
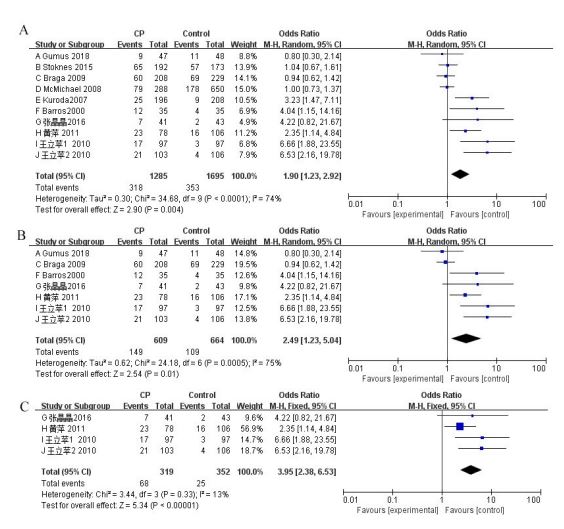
First, the meta-analysis of the APOE alleles and the CP risk was conducted. Overall, 10 studies were used to evaluate the effect of APOE alleles on CP risk [16,17,20-27]. Comparing the presence of ε2 vs. ε3 alleles within CP patients, as well as the control group, indicated heterogeneity between studies (P=0.01, χ² =21.37, I² =58%, Figure 2A). The random effects model was adopted. The findings showed that the existence of the ε2 allele conferred a risk of CP (P=0.04, OR 1.41, 95% CI 1.01 to 1.96, Figure 2A). Moreover, the presence of ε4 vs. ε3 alleles between CP patients and control groups was estimated. Because of the heterogeneity among the studies (P<0.00001, χ2=41.01, I² =78%, Figure 3A), the random effects model was used. The meta-analysis showed that there was a significant positive correlation between the ε4 allele and CP risk (P<0.001, OR 2.05, 95% CI 1.40 to 2.99, Figure 3A). Moreover, the pooled data supported the result that E4 carriers showed significantly increased CP risk, contrasted with those with the E3/3 genotype (P=0.004, OR 1.90, 95% CI 1.23 to 2.92, Figure 4A). The random effects model was adopted due to heterogeneity across the 10 studies (P<0.0001, χ² =34.68, I² =74%, Figure 4A). The results of dominant and recessive models for contrasts of E4, E3, and E2 genotypes are shown in Table 2. To further address the heterogeneity, we removed studies that showed a substantial departure from the HWE among controls. This fixed effects model was then applied because the heterogeneity was not significant among the pooled 7 studies (I² =44%, Figure 2B), and the meta-analysis showed that there was a significant positive correlation between the ε2 allele and CP risk (P=0.001, OR 1.63, 95% CI 1.21 to 2.19, Figure 2B)[16,20,23-27].
Figure 1:Flow diagram of the study selection process. CNKI, Chinese National Knowledge Infrastructure.
Figure 2:Forest plots describing the association of APOE polymorphism with cerebral palsy (CP) (ε2 allele versus ε3 allele). A: Overall analyses; B: Overall analyses (PHWE>0.05); C: Chinese subgroups analyses.
| ID | Study | Year | Population | Study type | Types of CP | Source of controls | genotyping methods | Sample size | NOS score | |
|---|---|---|---|---|---|---|---|---|---|---|
| CP | Control | |||||||||
| 1 | Gumus et al16 | 2018 | Anatolian | case–control | spastic (unilateral, bilateral)/dyskinetic/ ataxic/ unclassified CP | population-based | Real-time PCR | 78 | 60 | 7 |
| 2 | Stoknes et al17 | 2015 | Norse | case-parent triads | spastic (unilateral, bilateral)/dyskinetic/ ataxic/ unclassified CP | siblings | / | 295 | 256 | 4 |
| 3 | Xu et al18 | 2014 | Chinese | case–control | spastic/ataxic/ dyskinetic/ mixed/ hypotonic/ unclassified CP | population-based | MassARRAY | 350 | 242 | 9 |
| 4 | O’Callaghan et al19 | 2012 | Caucasian | case–control | hemiplegia/ diplegia/ quadriplegia/ other CP types | population-based | MassARRAY | 587 | 1154 | 9 |
| 5 | Braga et al20 | 2009 | Brazilian | cross-sectional | Spastic CP | hospital-based | Real-time PCR | 243 | 243 | 6 |
| 6 | McMichael et al21 | 2008 | Caucasian | case–control | diplegia/ hemiplegia/ quadriplegia, and all types CP | hospital-based | PCR—RFLP | 342 | 773 | 8 |
| 7 | Kuroda et al22 | 2007 | American | Cross-sectional | spastic CP | population-based | PCR—RFLP | 209 | 209 | 7 |
| 8 | Barros et al23 | 2000 | brazilian | case–control | mild or moderate CP | population-based | PCR—RFLP | 40 | 40 | 4 |
| 9 | 张晶晶et al24 | 2016 | Chinese | case–control | unclassified CP | population-based | PCR—RFLP | 50 | 51 | 9 |
| 10 | 黄萍et al25 | 2011 | Chinese | case–control | unclassified CP | population-based | PCR—RFLP | 83 | 120 | 9 |
| 11 | 王立苹1 et al26 | 2010 | Chinese | case–control | spastic CP | population-based | PCR—RFLP | 110 | 110 | 8 |
| 12 | 王立苹2 et al27 | 2010 | Chinese | case–control | unclassified CP | population-based | PCR—RFLP | 120 | 120 | 9 |
| a Not including overlapping data; NA, not available; CP, cerebral palsy; HWE, Hardy-Weinberg Equilibrium; RFLP, Restriction Fragment Length Polymorphism; | ||||||||||
Table 1: Characteristics of studies investigating the association of APOE polymorphisms with cerebral palsy.
APOE polymorphisms and CP susceptibility in Chinese subgroups
We also researched the subgroup of Chinese individuals because we involved four Chinese studies that had never appeared in other meta-analyses. In this paper, four studies of the ε4 vs. ε3 alleles were carried out [24-27]. The summary of the data supported a significant increase in the CP risk in individuals with ε4 alleles compared with that in those with ε3 alleles (P<0.00001, OR 3.70, 95% CI 2.37 to 5.78, Figure 3C). Because there was no heterogeneity between studies (I² = 9%, Figure 3C), a fixed effects model was then applied. We found that compared with those with ε4 alleles, individuals with ε2 alleles did not have a risk for CP development in the Chinese population (P=0.69, OR 1.09, 95% CI 0.72 to 1.65, Figure 2C). In addition, the summary data showed that those who were E4 carriers had a high risk of developing CP compared with individuals with the E3/3 genotype (P<0.00001, OR 3.95, 95% CI 2.38 to 6.53, Figure 4C).Because there was no heterogeneity between studies (I² =13%, Figure 4C), the fixed effects model was used. Table 2 shows the results comparing the dominant and recessive models of E4, E3 and E2 genotypes.
Figure 3:Forest plots describing the association of APOE polymorphism with cerebral palsy (CP) (ε4 allele versus ε3 allele). A: Overall analyses; B: Overall analyses (PHWE>0.05); C: Chinese subgroups analyses.
Figure 4:Forest plots describing the association of APOE polymorphism with cerebral palsy (CP) (E4 carriers versus E3/3 genotypes). A: Overall analyses; B: Overall analyses (PHWE>0.05); C: Chinese subgroups analyses.
Evaluation of publication Bias
First, Begg’s funnel plots were used to evaluate publication bias. Asymmetry and publication bias shown on funnel plots were evaluated by Egger’s test (Table 3). We found that comparisons of both ε4 vs ε3 alleles and E4 carriers vs E3/3 genotypes showed evidence of publication bias (P<0.05 for both Begg’s test and Egger’s test). In contrast, there was a significant deviation for both comparisons of ε2 vs ε3 alleles and E2 carriers vs E3/3 genotypes (P>0.05 for both Begg’s test and Egger’s test) (Figure S1A-D). Because of this result, we used the trim and fill method for sensitivity analysis, which conservatively presupposes hypothetical negative unpublished studies to reflect a positive study leading to the asymmetry in the funnel diagram [35]. The collected analysis incorporating the hypothetical studies continued to suggest that both APOE ε4 and E4 carriers act as risk factors for CP (Figure S1E-H).
Discussion
This is the first time a meta-analysis has been carried out to research the association between APOE polymorphisms and CP risk. In this meta-analysis, 10 qualified studies were included, of which 5 studies showed that the APOE ε4 allele is a risk factor [23-27], 1 study indicated that the APOE ε2 allele is a risk factor [22], 2 studies indicated that both APOE ε2 and ε4 alleles act as risk factors [16,22], and 1 study suggested that APOE allelic and genotypic frequencies did not differ between patients and controls [21]. To reconcile these contradictory findings with a larger sample size, we have conducted a systematic review of the published studies. In this meta-analysis, a total of 1570 CP patients and 1982 healthy subjects were used to assess the relationship between APOE polymorphism and CP. This meta-analysis indicated that individuals carrying the APOE ε4 allele, especially in the Chinese population, had an increased risk of CP (Figure 3A and 3C). We also found a highly significant association between E4 carriers and CP development risk, especially in the Chinese population (Figure 4A and 4C).
| Polymorphisms | Comparisons | Population | Number of studies | Test of association | Test of heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Z | P value | Model | χ² | P value | I² (%) | ||||
| ε2 | ε2 vs ε3 alleles | Overall† | 10 | 1.41 [1.01, 1.96] | 2.01 | 0.04 | R | 21.37 | 0.01 | 58% |
| Overall† | 7 | 1.63 [1.21, 2.19] | 3.21 | 0.001 | F | 10.75 | 0.10 | 44% | ||
| Chinese ‡ | 4 | 1.09 [0.72, 1.65] | 0.40 | 0.69 | F | 2.52 | 0.47 | 0% | ||
| E2 carriers vs E3/3 | Overall* | 10 | 1.16 [0.92, 1.46] | 1.22 | 0.22 | F | 15.15 | 0.09 | 41% | |
| Overall† | 7 | 1.17 [0.83, 1.66] | 0.88 | 0.38 | F | 7.93 | 0.24 | 24% | ||
| Chinese ‡ | 4 | 0.95 [0.59, 1.53] | 0.20 | 0.84 | F | 2.02 | 0.57 | 0% | E2/2 vs E2/3+E3/3 | Overall* | 10 | 1.13 [0.60, 2.12] | 0.37 | 0.71 | F | 3.24 | 0.66 | 0% |
| Overall† | 7 | 2.32 [0.57, 9.39] | 1.18 | 0.24 | F | 1.65 | 0.65 | 0% | ||
| Chinese ‡ | 4 | 1.66 [0.33, 8.50] | 0.61 | 0.54 | F | 1.24 | 0.54 | 0% | ||
| E2/2 vs E3/3 | Overall* | 10 | 1.12 [0.59, 2.11] | 0.34 | 0.73 | F | 3.08 | 0.69 | 0% | |
| Overall† | 7 | 2.25 [0.55, 9.16] | 1.13 | 0.26 | F | 1.56 | 0.67 | 0% | ||
| Chinese ‡ | 4 | 1.68 [0.33, 8.58] | 0.62 | 0.53 | F | 1.24 | 0.54 | 0% | ||
| ε4 | ε4 vs ε3 alleles | Overall* | 10 | 2.05 [1.40, 2.99] | 3.71 | 0.0002 | R | 41.01 | <0.00001 | 78% |
| Overall† | 7 | 2.78 [1.51, 5.09] | 3.30 | 0.0010 | R | 25.29 | 0.003 | 76% | ||
| Chinese ‡ | 4 | 3.70 [2.37, 5.78] | 5.75 | <0.00001 | F | 3.30 | 0.35 | 9% | ||
| E4 carriers vs E3/3 | Overall* | 10 | 1.90 [1.23, 2.92] | 2.90 | 0.004 | R | 34.68 | <0.0001 | 74% | |
| Overall† | 7 | 2.49 [1.23, 5.04] | 2.54 | 0.01 | R | 24.18 | 0.0005 | 75% | ||
| Chinese ‡ | 4 | 3.95 [2.38, 6.53] | 5.34 | <0.00001 | F | 3.44 | 0.33 | 13% | ||
| E4/4 vs E3/3+E3/4 | Overall* | 10 | 1.22 [0.73, 2.02] | 0.76 | 0.45 | F | 3.06 | 0.93 | 0% | |
| Overall† | 7 | 1.22 [0.50, 2.95] | 0.44 | 0.66 | F | 1.97 | 0.85 | 0% | ||
| Chinese ‡ | 4 | 2.93 [0.56, 15.35] | 1.27 | 0.20 | F | 0.00 | 1.00 | 0% | ||
| E4/4 vs E3/3 | Overall* | 10 | 1.27 [0.76, 2.10] | 0.91 | 0.36 | F | 3.42 | 0.91 | 0% | |
| Overall † | 7 | 1.32 [0.54, 3.19] | 0.61 | 0.54 | F | 2.16 | 0.83 | 0% | ||
| Chinese ‡ | 4 | 3.46 [0.66, 18.18] | 1.47 | 0.14 | F | 0.00 | 1.00 | 0% | ||
| E2 carrier include E2/2 and E2/3; E4 carrier include E3/4 and E4/4; Overall*, Overall analyses; Overall†, Overall analyses (PHWE>0.05); Chinese ‡, Chinese subgroups analyses; OR, odds ratio; R, random-effects model; F, fixed-effects model | ||||||||||
Table 2: Meta-analysis of the association of APOE polymorphisms and cerebral palsy.
| Publication bias by Egger’s test | |||||
|---|---|---|---|---|---|
| Variables | Coefficient | SE | Z | P Value | 95% CI |
| ε2 vs ε3 alleles | 4.983226 | 1.93 | 2.575458 | 0.089 | -.9557911 to 10.92224 |
| E2 carriers vs E3/3 | 4.906202 | 3.225086 | 1.52 | 0.167 | 2.530861 to 12.34326 |
| ε4 vs ε3 alleles | 8.115601 | 1.398786 | 5.80 | 0.000 | 4.889996 to 11.34121 |
| E4 carriers vs E3/3 | 7.736085 | 1.921866 | 4.03 | 0.004 | 3.304254 to 12.16792 |
Table 3: Publication bias of APOE polymorphisms and the risk of CP.
The APOE ε2 allele also appeared to be related to an increased risk of CP, but not appeared in the Chinese population (Figure 2A and 2C). However, in addition to E4 carriers, we found no significant associations between other APOE polymorphisms and the risk of CP development. The results of our study suggested that APOE ε4 is an important genetic risk factor for the development of CP.
Apolipoprotein E is one of the main apolipoproteins in the central neuronal system that plays an important role in neurobiology. Between the APOE ԑ4 allele and CP, the existence of an association has been defined in many studies [16,22,23-27]. Disturbances in neurobehavioral functions and the brain healing process, along with reduced ischaemia tolerance, have all been shown to be related to the possession of the APOE ԑ4 allele in a number of studies [17,36]. Interestingly, against poor prognosis and unfavourable clinical outcomes stemming from the ԑ4 allele, some studies suggest that having the APOE ԑ3 allele renders a favourable response to traumatic and hypoxic injury in the developing brain [37]. A meta-analysis of 2,000 adults aged 45-89 years found that APOE ε4 resulted in poor executive function in cognitive assessment. It is suggested that the efficiency of nerve cell repair is low in allele ԑ4 carriers [38]. Combined with the results of our analysis, we concluded that the risk of CP was significantly increased in ε4 allele individuals.
The relationship between the APOE ε2 allele and CP is contradictory. BRAGA et al found that the frequency distribution of the ε2 allele in individuals with CP was significantly higher than that in the control group [20]. Another study conducted by McMicheal reported an association between the ԑ2 allele and low birth weight, as well as prematurity [21]. Our data show that the APOE ε2 allele increases the risk of CP slightly in multi-ethnic samples, but this trend is not obvious in the Chinese population. The different ethnicities, races and environments of the sample population might be part of the reason why the literature produces contradictory results regarding the relationship between the ε2 allele and CP.
Some limitations of this research should be discussed. First, the meta-analysis was based on unadjusted data due to a lack of individual original data, and a more accurate analysis of hierarchical environmental factors or clinical manifestations was not carried out. Second, in some studies, the distribution of genotypes in the control group did not align with the HWE, which may affect the validity of the conclusion. Third, funnel plot analysis showed some asymmetrical phenomena, indicating the existence of publication bias. Sensitivity analysis was carried out by the trim and fill method, and the results show that this association is not an artefact of unpublished negative studies (Figure S1). However, this approach does not completely rule out this possibility. Fourth, although we detected an association between APOE genetic polymorphisms (ε2 vs. ε3 alleles; ε4 vs. ε3 alleles; E4 carriers vs. E3/3 genotypes) and CP, the result should be approached with caution because the number of participants was small.
Conclusion
In summary, the pooled data indicate a high correlation between APOE polymorphisms and CP. In contrast to individuals carrying the APOE ε3 allele, the risk of CP was significantly increased in individuals carrying the ε4 allele. In addition, compared with individuals with the APOE E3/3 genotype, E4 carriers have a significantly increased risk of CP. Because of the small number studies, further welldesigned studies are still warranted to confirm whether the APOE ε2 allele increases susceptibility to CP. Additionally, the mechanism of apolipoprotein E involvement in CP is not clear and needs to be further studied.
Authors’Contribution
HC: Provided contributions to the design of study, extraction of data, analysis and interpretation of data. Drafted and critically revised the manuscript, and approved of the final version. HY and YC: Provided contributions to the extraction and analysis of data. CC: Provided contributions to the revision of the manuscript and approved of the final version. XZ: Provided contributions to the conception and design of study, extraction of data, analysis and interpretation of data. Revised the manuscript critically, and approved of the final version. All authors read and approved the final manuscript.
Funding
This work was supported by the Shaanxi Natural Science Basic Research Project, China (NO.2020JQ-958); and Key Science and Technology Program of Shaanxi Province, China (NO.2017SF-062). The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Acknowledgement
Not applicable.
Compliance with Ethical Standards
Conflict of interests
The authors declare that they have no conflict of interests.
Statement of compliance with standards of research involving humans as subjects
Statement of compliance with standards of research involving humans as subjects. All procedures performed in the study with the participation of people corresponded to the ethical standards of the Commission on Bioethics , and the Helsinki Declaration 1964 with its subsequent amendments or comparable ethical norms. All subjects of those study gave their voluntary consent to participate in that study and signed their informed consent.
References
Christensen D, Van Naarden Braun K, Doernberg NS, Maenner MJ, et al. (2014) Prevalence of cerebral palsy, co-occurring autism spectrum disorders, and motor functioning - Autism and Developmental Disabilities Monitoring Network, USA, 2008. Developmental medicine and child neurology 56: 59-65.[ Ref ]
Reid SM, Meehan E, McIntyre S, Goldsmith S, Badawi N (2016) Temporal trends in cerebral palsy by impairment severity and birth gestation. Developmental medicine and child neurology 58S2: 25-35.[ Ref ]
Mei C, Reilly S, Reddihough D, Mensah F, Pennington L (2016) Language outcomes of children with cerebral palsy aged 5 years and 6 years: a population-based study. Developmental medicine and child neurology 58: 605-611.[ Ref ]
Levy SE, Giarelli E, Lee LC, Schieve LA, Kirby RS (2010) Autism spectrum disorder and co-occurring developmental, psychiatric, and medical conditions among children in multiple populations of the United States. Journal of developmental and behavioral pediatrics. JDBP 31: 267-275.[ Ref ]
Delobel-Ayoub M, Klapouszczak D, van Bakel MME (2017) Prevalence and characteristics of autism spectrum disorders in children with cerebral palsy. Developmental Medicine & Child Neurology 59: 738-742.[ Ref ]
Korzeniewski SJ, Slaughter J, Lenski M, Haak P, Paneth N (2018) The complex aetiology of cerebral palsy. Nature reviews Neurology 14: 528- 543.[ Ref ]
Li XJ, Qiu HB, Jiang ZM, Pang W, Guo J (2018) Epidemiological characteristics of cerebral palsy in twelve province in China. Journal of Applied Clinical Pediatrics 33: 378-383.[ Ref ]
Kancherla V, Amendah DD, Grosse SD, Yeargin-Allsopp M, Van Naarden Braun K (2012) Medical expenditures attributable to cerebral palsy and intellectual disability among Medicaid-enrolled children. Res Dev Disabil 33: 832-840.[ Ref ]
Campagna J, Spilman P, Jagodzinska B (2018) A small molecule ApoE4- targeted therapeutic candidate that normalizes sirtuin 1 levels and improves cognition in an Alzheimer’s disease mouse model. Sci Rep 8: 17574.[ Ref ]
Lima D, Hacke ACM, Inaba J, Pessôa CA, Kerman K (2020) Electrochemical detection of specific interactions between apolipoprotein E isoforms and DNA sequences related to Alzheimer’s disease. Bioelectrochemistry 133: 107447.[ Ref ]
Theendakara V, Patent A, Peters Libeu CA, Philpot B, Flores S, et al. (2013) Neuroprotective Sirtuin ratio reversed by ApoE4. Proc Natl Acad Sci USA 110: 18303-18308.[ Ref ]
.Rall SC, Jr Weisgraber KH, Mahley RW (1982) Human apolipoprotein E. The complete amino acid sequence. The Journal of biological chemistry 257: 4171-4178.[ Ref ]
Weisgraber KH, Rall SC, Mahley RW (1981) Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. The Journal of biological chemistry 256: 9077-9083.[ Ref ]
Bretsky P, Guralnik JM, Launer L, Albert M, Seeman TE (2003) The role of APOE-epsilon4 in longitudinal cognitive decline: MacArthur Studies of Successful Aging. Neurology 60: 1077-1081.[ Ref ]
Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261: 921-923.[ Ref ]
Gumus E, Aras BD, Cilingir O, Yarar C, Carman KB (2018) Apolipoprotein E allelic variants and cerebral palsy. The Turkish journal of pediatrics 60: 361-371.[ Ref ]
Stoknes M, Lien E, Andersen GL, Bao Y, Blackman JA (2015) Child apolipoprotein E gene variants and risk of cerebral palsy: estimation from case-parent triads. European journal of paediatric neurology: EJPN : official journal of the European Paediatric Neurology Society 19: 286- 291[ Ref ]
Xu Y, Wang H, Sun Y, Shang Q, Chen M (2014) The association of apolipoprotein E gene polymorphisms with cerebral palsy in Chinese infants. Molecular genetics and genomics: MGG 289: 411-416.[ Ref ]
O'Callaghan ME, Maclennan AH, Gibson CS, McMichael GL, Haan EA (2012) Fetal and maternal candidate single nucleotide polymorphism associations with cerebral palsy: a case-control study. Pediatrics 129: e414-423.[ Ref ]
Blackman JA (2010) Apolipoprotein E genotype and cerebral palsy. Developmental medicine and child neurology 52: 666-671.[ Ref ]
.McMichael GL, Gibson CS, Goldwater PN, Haan EA, Priest K (2008) Association between Apolipoprotein E genotype and cerebral palsy is not confirmed in a Caucasian population. Hum Genet 124: 411-416.[ Ref ]
Kuroda MM, Weck ME, Sarwark JF, Hamidullah A, Wainwright MS (2007) Association of apolipoprotein E genotype and cerebral palsy in children. Pediatrics 119: 306-313.[ Ref ]
Meirelles Kalil Pessoa de B, Rodrigues CJ, de Barros TE, Bevilacqua RG (2000) Presence of apolipoprotein E epsilon4 allele in cerebral palsy. Journal of pediatric orthopedics 20: 786-789.[ Ref ]
Zhang JJ (2016) Study on the relationship between apolipoprotein E gene polymorphism and Xinjiang Uygur children with cerebral palsy. Journal of Clinical Medical Literature (Electronic Edition) 3: 9733-9736.[ Ref ]
Huang P (2011) Study on the Relationship between Apoliprotein E Gene Polymorphism and Children Cerebral Palsy [D]. Guangxi Medical University[ Ref ]
Wang L, Li X, Zhu J (2010) Preliminary investigation of association between ApoE genotype and spastic cerebral palsy in children. Chinese Journal of Birth Health & Heredity 5: 10.[ Ref ]
Wang LP, Li XJ, Zhang H, Sun QF (2010) Association between ApoE Genotype and Cerebral Palsy Combined with Mental Retardation. Chinese Journal of Rehabilitation 25: 259-261.[ Ref ]
Zintzaras E, Ioannidis JP (2005) Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol 28: 123-137.[ Ref ]
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses BMJ 327: 557-560.[ Ref ]
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Statistics in Medicine 21: 1539-1558.[ Ref ]
Dersimonian R, Laird N (1986) Meta-analysis in clinical trials. Controlled Clinical Trials 7: 177-188.[ Ref ]
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute 22: 719-748.[ Ref ]
Seagroatt V, Stratton I (1998) Bias in meta-analysis detected by a simple, graphical test. Test had 10% false positive rate, BMJ 316: 470.[ Ref ]
Begg CB, Berlin JA (1988) Publication bias: a problem in interpreting medical data. Journal of the Royal Statistical Society: Series A (Statistics in Society) 151: 419-445.[ Ref ]
Duval S, Tweedie R (2000) Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 56: 455-463.[ Ref ]
Kim J, Basak JM, Holtzman DM (2009) The role of apolipoprotein E in Alzheimer’s disease. Neuron 63: 287-303.[ Ref ]
Lo TY, Jones PA, Chambers IR, Beattie TF, Forsyth R (2009) Modulating effect of apolipoprotein E polymorphisms on secondary brain insult and outcome after childhood brain trauma. Child’s nervous system: ChNS : official journal of the International Society for Pediatric Neurosurgery 25: 47-54.[ Ref ]
Mahley RW, Weisgraber KH, Huang Y (2006) Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc Natl Acad Sci USA 103: 5644-5651.[ Ref ]