Journal Name: Journal of Pediatrics and Infants
Article Type: Research
Received date: 11 March 2019
Accepted date: 22 March ,2019
Published date: 28 March, 2019
Citation: Raj AY, Ahmed F (2019) Emerging Multi Drug Resistant Bacterial Strain in Neonatal Intensive (NICU) in a Developing Country-Bangladesh. J Pediat Infants Vol: 2, Issu: 1 (01-07).
Copyright: © 2019 Raj AY. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Introduction: Neonatal sepsis is one of the major causes for mortality and morbidity among newborn. Bacterial pathogens and antibiotic sensitivity patterns are different in hospitals of each country. The objective of the present study was to identify the spectrum of bacterial isolates causing septicemia in neonates and to determine their antimicrobial sensitivity pattern.
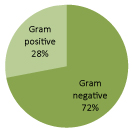
Results: Out of 171 babies with positive septic screening, 82 were culture positive samples, gram positive and gram negative organisms were 28 % and 72 % respectively. Acinetobacter remained the predominant isolate (n=25,30.5%) followed by Coagulase negative Staphylococcus (N=16, 19.5%). Most of the gram positive isolates exhibited higher resistance to penicillin, cephalosporin, macrolides, gentamycin in aminoglycosides group and quinolones. Gram-positive isolates had sensitivity of 100% to chloramphenicol, vancomycin, linezolid followed by rifampicin 84%. In comparison to other antibiotics, sensitivity to these four medicines was significant statistically (P < 0.05). On the other hand, most of the gram negative organisms showed resistance to aminoglycosides and cephalosporin; about two third showed resistant to meropenem, quinolones and zosyn. Best overall sensitivity among Gram-negative isolates was to Polymixin B (100%) and Minocycline (97%), followed by Colistin (83%) which was also significant statistically (P < 0.05).
Conclusion: Our study highlights the higher resistance towards commonly used antimicrobials and necessity for periodic survey of etiological agents and their antibiotic susceptibility patterns for the timely alarm of such type of problems.
Keywords
Antimicrobial Resistance, Antibiotics, Neonatal Septicemia.
Abstract
Introduction: Neonatal sepsis is one of the major causes for mortality and morbidity among newborn. Bacterial pathogens and antibiotic sensitivity patterns are different in hospitals of each country. The objective of the present study was to identify the spectrum of bacterial isolates causing septicemia in neonates and to determine their antimicrobial sensitivity pattern.
Results: Out of 171 babies with positive septic screening, 82 were culture positive samples, gram positive and gram negative organisms were 28 % and 72 % respectively. Acinetobacter remained the predominant isolate (n=25,30.5%) followed by Coagulase negative Staphylococcus (N=16, 19.5%). Most of the gram positive isolates exhibited higher resistance to penicillin, cephalosporin, macrolides, gentamycin in aminoglycosides group and quinolones. Gram-positive isolates had sensitivity of 100% to chloramphenicol, vancomycin, linezolid followed by rifampicin 84%. In comparison to other antibiotics, sensitivity to these four medicines was significant statistically (P < 0.05). On the other hand, most of the gram negative organisms showed resistance to aminoglycosides and cephalosporin; about two third showed resistant to meropenem, quinolones and zosyn. Best overall sensitivity among Gram-negative isolates was to Polymixin B (100%) and Minocycline (97%), followed by Colistin (83%) which was also significant statistically (P < 0.05).
Conclusion: Our study highlights the higher resistance towards commonly used antimicrobials and necessity for periodic survey of etiological agents and their antibiotic susceptibility patterns for the timely alarm of such type of problems.
Keywords
Antimicrobial Resistance, Antibiotics, Neonatal Septicemia.
Introduction
Neonatal sepsis is a major health problem worldwide, with the global prevalence of (1-10) per 1000 live births. It is estimated that up to 20% of neonates develop sepsis and approximately 1% die of sepsis related causes [1]. In spite of significant advances of neonatal care and the development of broad spectrum antibiotic, sepsis remains a leading cause of morbidity and mortality in newborn, specially, those born prematurely and with low birth weight [2]. Since the aim of empirical therapy is to target the most likely infectious organism(s), it is crucial for neonatal unit to survey the profile of causative organism and their susceptibility in order to ensure effective antimicrobial treatment [3]. Statistical data from different countries shows differences in the pattern, risk factors, incidence and antimicrobial sensitivities of pathogens and mortality from that of developed countries [3,4]. In the Europe and North America, group B streptococcus and E-coli contribute to 70%-75% of cases of neonatal septicemia [3-5]. In most of developing countries, Gram negative organisms remain the major cause of neonatal sepsis, particularly early onset sepsis [3-5].
As neonatal sepsis is a life threatening emergency and delay in diagnosis and treatment may have adverse consequences, surveillances are needed to identify antibiotic sensitivity pattern of common pathogens [6].
Materials and Methods
The study was conducted retrospectively in the in the NICU at Square Hospital Ltd. which is a tertiary care center at Dhaka, Bangladesh, from January 2012 and June 2016. Out of 171 positive cases of septic screening, all cultures positive cases were included in the study and skin commensals, contaminates and fungal pathogens were excluded. After collection of samples (2-3 ml blood for blood culture, urine for culture, umbilical swab, conjunctival swab, culture of tip of endotracheal tube, culture of aspirate from endotracheal tube) with all aseptic precaution, culture bottles were transported immediately to the Microbiology Laboratory and were processed as per standard microbiological techniques and the isolates were identified. All positive culture reports were checked, verified and analyzed. The sensitivity and resistance pattern of various antibiotics against the isolated pathogens were also noted. Data was expressed as P -value and percentages. was used for data was analyzed using. The obtained data was statistically analyzed using Statistical Package for Social Sciences (SPSS) version 16 Fisher’s exact test using an r × c exact contingency table. A P value of less than 0.05 was considered to be significant. Approval for the study was taken from the ethical committee of the hospital.
Results
All the culture reports were checked and verified and of them eighty two culture positive reports were finally analyzed. The positive yield of blood, urine, eye swab, tip of endotracheal tube and aspirate from endotracheal tube cultures were 73%, 1.2%, 1.2%, 18.29% and 3.66% respectively (Table 1).
Table 1: Source of various isolates causing neonatal infection.
| Source of sample | Number | Percentage |
|---|---|---|
| blood culture | 60 | 73 |
| Culture of tip of endotracheal tube | 15 | 18.29 |
| Culture of aspirate from endotracheal tube | 3 | 3.66 |
| Culture of umbilical swab | 2 | 2.44 |
| Culture of conjunctival swab | 1 | 1.2 |
| Urine culture | 1 | 1.2 |
Out of these 82 bacterial isolates, 59 (72%) were Gram-negative organisms, 23 (28%) were Gram-positive organisms. The organisms causing sepsis are shown in Figure 1.
Figure 1: Organism responsible for neonatal sepsis.
According to table 2, Acinetobacter spp. (30.49%) are the commonest organism responsible for infection in neonate followed by Coagulase negative staphylococci (19.5%
Table 2: Percentage of various isolates.
| Organism | No. of isolates | Percentage |
|---|---|---|
| Gram-positive organisms | ||
| Coagulase negative staphylococci | 16 | 19.5 |
| Streptococcus pneumonia | 3 | 3.66 |
| Enterococcus faecium | 4 | 4.88 |
| Gram-negative organisms | ||
| Acinetobacter spp | 25 | 30.49 |
| Klebsiella species | 14 | 17.07 |
| Enterobacter | 5 | 6.10 |
| E.coli | 1 | 1.21 |
| Burkholderia cepacia | 4 | 4.88 |
| Chryseobacterium | 2 | 2.44 |
| Stenotrophomonas Maltophilia | 7 | 8.54 |
| Pseudomonas spp | 1 | 1.21 |
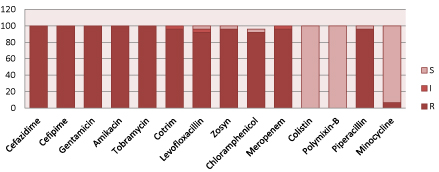
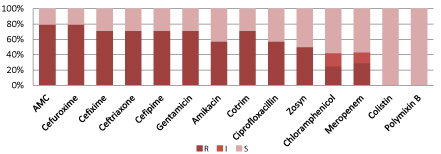
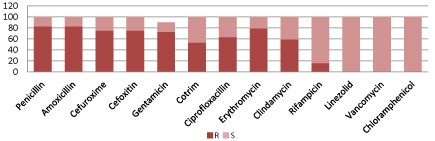
According to figure 2, Acinetobacter spp. are resistant to commonly used antibiotics including meropenem and amikacin but 100% sensitive to Colistin and polymixin-B.
Figure 2: Resistant pattern of Acinetobacter spp.
(S=Sensitive, I=intermediate sensitive, R=Resistant, Zosyn=Piperacillin/Tazobactam, Cotrim: Trimethoprim/Sulfamethoxazole).
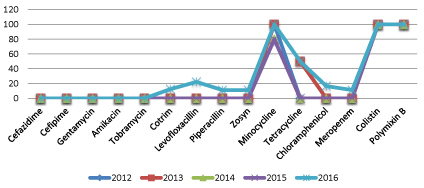
Surprisingly, sensitivity pattern for Acinetobacter spp. have not been changed significantly over last four and half years (Figure 3).
Figure 3: Percentage of Antibiotic sensitivity pattern for Acinetibacter spp. over four and half years.
Figure 4 shows, Stenotrophomonas Maltophilia are resistant to commonly used antibiotics including meropenem and colistin but 100% sensitive to Minocycline and Zosyn (Piperacillin and tazobactam)
Figure 4: Resistant pattern for Stenotrophomonas maltrophilia.
(S=Sensitive, I=intermediate sensitive, R=Resistant, Zosyn=Piperacillin/Tazobactam, Cotrim: Trimethoprim/Sulfamethoxazole).
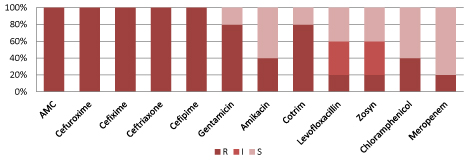
According to figure 5, Klebsiella species. are resistant to commonly used antibiotics but 100% sensitive to Colistin and polymixin-B.
Figure 5: Resistant pattern for Klebsiella spp.
(S=Sensitive, I=intermediate sensitive, R=Resistant, Zosyn=Piperacillin/Tazobactam, Cotrim: Trimethoprim/Sulfamethoxazole).
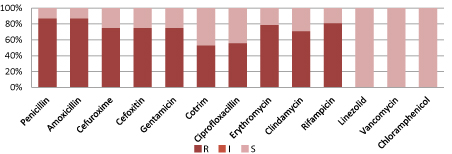
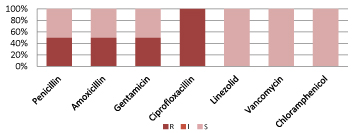
Figure 6 shows, Coagulase negative Staphylococcus are resistant to commonly used antibiotics but 100% sensitive to Linezolid, Vancomycin and Chloramphenicol.
Figure 6: Resistant pattern for CoNS.
(S=Sensitive, I=intermediate sensitive, R=Resistant; CoNS=Coagulase negative Staphylococcus, Zosyn=Piperacillin/Tazobactam,
Cotrim: Trimethoprim/Sulfamethoxazole).
Figure 7 shows, Enterobacter spp. are resistant to commonly used antibiotics including third generation cephalosporin, 80% sensitive to Meropenem. It is 40% totally sensitive, 40% intermediately sensitive and 20% resistant to Zosyn and Levofloxacillin.
Figure 7: Resistant pattern for Enterobacter spp.
(S=Sensitive, I=intermediate sensitive, R=Resistant, AMC= Amoxicillin/Clavulanate, Zosyn=Piperacillin/Tazobactam, Cotrim:
Trimethoprim/Sulfamethoxazole).
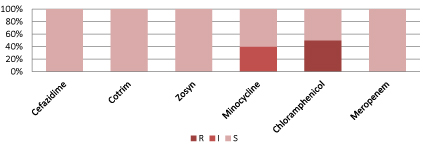
Figure 8 shows Burkholderia cepacia is 100% sensitive to ceftazidime, cotrim, Zosyn and Meropenem.
Figure 8: Resistant pattern for Burkholderia cepacia.
(S=Sensitive, I=intermediate sensitive, R=Resistant, Zosyn=Piperacillin/Tazobactam, Cotrim: Trimethoprim/Sulfamethoxazole).
Figure 9 shows, Stenotrophomonas Maltophilia are resistant to commonly used antibiotics but 100% sensitive to Linezolid, Vancomycin and Chloramphenicol.
Figure 9: Resistant pattern for Entyerococcus faecium.
(S=Sensitive, I=intermediate sensitive, R=Resistant, Zosyn=Piperacillin/Tazobactam, Cotrim: Trimethoprim/Sulfamethoxazole).
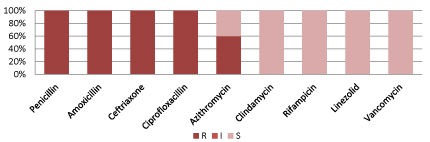
Figure 10 shows, Streptococcus pneumoniae resistant to commonly used antibiotics but 100% sensitive to Clindamycin, Rifampicin, Linezolid and Vancomycin.
Figure 10: Resistant pattern for Entyerococcus faecium.
(S=Sensitive, I=intermediate sensitive, R=Resistant, Zosyn=Piperacillin/Tazobactam, Cotrim: Trimethoprim/Sulfamethoxazole).
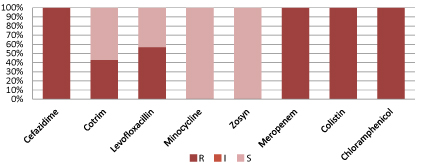
Figure 11 showing overall sensitivity pattern of gram positive organism. Most of the gram positive isolates exhibited higher resistance to penicillin, cephalosporin, macrolides, gentamicin and quinolones. Susceptibility to commonly used was found to vancomycin (100%), chloramphenicol (100%), rifampicin (84%) and Linezolid (100%). In comparison to other commonly used antibiotics, sensitivity to these four medicines was statistically significant (P < 0.05).
Figure 11: Antibiotic Resistance pattern for gram positive organism.
(P < 0.05; Fisher’s exact test)
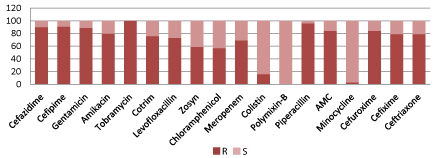
Figure 12 showing overall sensitivity pattern of gram negative organism. Most of the gram negative bacteria showed resistance to cephalosporin, aminoglycosides; about two third showed resistant to meropenem, zosyn and quinolones. Best overall sensitivity among Gram-negative isolates was to Polymixin B (100%) and Minocycline (97%), followed by Colistin (83%). In comparison to other commonly used antibiotics, sensitivity to these three medicines was statistically significant (P < 0.05).
Figure 12: Antibiotic resistant pRatternS of gram negative organism.
Discussion
Neonatal sepsis remains a major clinical problem in neonatology with high morbidity and mortality rate specially in developing countries like Bangladesh [7]. The knowledge of etiological organisms as well as their antimicrobial sensitivity profile is necessary for effective therapeutic intervention in neonatal sepsis [8]. Multiple drug resistance of the causative organisms of sepsis is a rapidly emerging and potentially disastrous problem because infection with resistant organisms has been associated with treatment failure, higher mortality and morbidity, increased health costs and prolonged hospitalization [9].
In the present study Gram negative organisms were the major cause of neonatal sepsis about 72% and gram positive organisms about 28%. A recent study conducted by Muley V A et al. [1]. revealed 70.8% neonatal septicemia caused by Gram-negative organism. Almost similar results were reported by Pooja R et al. where Gram negative and gram positive organisms were isolated in 79.9% and 18.17% cases respectively [10] reported that Gram-positive organisms caused 73% of bacterial sepsis but Gram-negative organisms were responsible for the highest mortality rate [11].
The resistant pattern of the organisms can be minimized by periodic epidemiological surveys of etiological agents and their antibiotic sensitivity patterns leading to recognition of the most frequently encountered pathogens in a particular geographical area [12].
The organism causing neonatal sepsis in developing countries differ from those seen in developed countries. Overall, Gram-negative organisms are more common and are mainly represented by Acinetobacter, Klebsiella, Escherichia coli and Pseudomonas. Of the Gram- positive organisms, Staphylococcus aureus, CONS, Streptococcus pneumonia, and S. pyogenes are most commonly isolated [13]. In our study, we found Acinetobacter remained the predominant isolate (n=25, 30.5%) followed by Coagulase negative Staphylococcus (N=16, 19.5%) implicated in neonatal sepsis. [14,15] reported CoNS mainly responsible for neonatal sepsis and were sensitive to vancomycin only; [14,15] we found Coagulase negative Staphylococcus were100% sensitive to Linezolid, Vancomycin and Chloramphenicol.
The present study shows a high degree of resistant of gram negative organisms to commonly used antibiotics like aminoglycosides and cephalosporin; about two third cases showed resistant to meropenem, zosyn and quinolones. Similar association had also been found in many other studies [16,17]. According to [17] this multidrug resistance pattern in Gram- negative organisms might be due to the production of extended spectrum beta-lactamases by Gramnegative organisms [18].
In this study, among Gram-negative isolates, maximum sensitivity was observed to Polymixin B (100%) and Minocycline (97%), followed by Colistin (83%). While compared to other commonly used antibiotics, sensitivity to these three medicines was statistically significant (P < 0.05). Mustafa M et al. reported higher sensitivity to imipenem and linezolid which were statistically significant (P < 0.05), but they recommended not to use these medicines indiscriminately to prevent resistance to these drugs may develop [19].
It is difficult to compare antibiotic resistance between countries and among different periods in a same country because the epidemiology of neonatal sepsis is extremely variable. Few studies compare antibiotic susceptibility over time in the same unit, but where data are available, they show increasing resistance to commonly used antibiotics [13]. Surprisingly, in our study we found that antibiotic sensitivity pattern for Acinetobacter spp. have not been changed significantly over last four and half years (Figure 3).
As the most common isolates in our study were Acinetobacter spp and Coagulase negative Staphylococcus, some of these could be contaminants only. Since, we have not analyzed each blood culture positive cases in relation with gestational age, weight, number of days in the NICU, timing of blood cultures, we cannot clearly say how many are likely to be contaminants. The indication of blood culture and the clinical details of the babies with positive or negative bacterial isolates were not analyzed in this study.
Conclusion
With the given antibiotic susceptibility pattern, there is no single antibiotic among the first line or second line drugs that can be used empirically in neonates with suspected gram positive and gram negative sepsis. Continuous surveillance is needed to monitor the changing epidemiology of pathogens and antibiotic susceptibility. Strategies to overcome the increasing incidence of resistance to conventional drugs are required.
There are no references